
Functional prediction of human erythrocytic miR-451a on Plasmodium falciparum 3D7 transcriptome—an in-silico study
Introduction
Globally, an estimated 229 million malaria cases were recorded in 2019, in 87 malaria endemic countries including India. Among the five species, which mainly infect human beings complicated malaria is caused only by the parasitic protozoan Plasmodium falciparum (P. falciparum). P. falciparum is the most widespread malaria parasite not only in the African Region, but also in the South-East Asian Region (1). The virulent nature of P. falciparum contributes largely to malaria related deaths (2), in which India, accounts for about 86% of cases in the South-East Asia Region, including pregnant women and children (<5 years). Cytoadherence, sequestration and rosetting, like characteristics make it more dangerous (1,3).
In the case of malaria, to minimize transmission, risk of development of parasite resistance and adverse reactions to anti-malarial drugs, rapid and accurate diagnosis as well as identification of Plasmodium spp. is mandatory (4). The diagnostic tools have evolved from conventional microscopic observation of Giemsa-stained blood films to rapid diagnostic tests (RDTs), serology, fluorescence microscopy and molecular diagnostic tools like PCR, qPCR and loop-mediated isothermal amplification (LAMP) (5,6). Yet newer, improved diagnostics are an essential prerequisite to obtain easy-to-use low-cost tests, to enhance identification of the drug resistant profile of the patients. The emergence and spread of drug resistant strains of P. falciparum over a period, has contributed to the re-emergence of complicated malaria in the world (7,8). To overcome this problem, priority should be given to identify novel therapeutic approaches in global malaria research.
Since last, few years’ researchers have been focussing on small noncoding RNA families in infectious diseases to understand novel perceptions of host parasitic interactions. Small noncoding RNA families mainly microRNAs (miRNAs) have emerged as one of the key players in the stringent, highly coordinated control of gene expression (9,10). miRNA molecules of ~21–25 nucleotides in length, synthesized in the nucleus and with the help of ribonucleoprotein complexes like RNA Induced Silencing Complex (RISC) directed towards target mRNAs to facilitate binding at 5' UTR region on it are mainly involved in transcriptional suppression through mRNA cleavage (10,11). They have also been found to bind on 3' UTR regions, promoters and translated regions of DNA sequences (12). There are reports that miRNAs also affect rate of translation and activation of genes etc. (10). As reported in earlier studies miRNAs are also secreted extracellularly in form of exosomes or bound with proteins and are involved in cell-cell communications (13).
Erythrocytes cannot synthesize miRNAs via the canonical pathway as, they are enucleated. They may retain miRNAs populations since enucleation (14). In addition, it has been observed that mature erythrocytes are proved as potential repositories of miRNAs in the circulating system (15). miRNAs can affect erythropoiesis by down regulation of miR-221, -222, -150 and -155 from transition of reticulocytes to mature erythrocytes (16,17). miR-451, -144 are reported to be upregulated during maturation of erythrocytes by GATA-1 (14,15,18). miR-451 increases the activity of FOXO3—which is involved in the control of oxidative stress and promotes synthesis of antioxidants. miR-451 with miR-223 and let-7i are found to be elevated in Sickle Cell Anemia traits HbSS as well as in HbAS (19,20).
Protozoans have a slightly different mechanism with regard to the miRNA biogenesis pathway as compared to metazoans. It has been found that miRNA biogenesis is independent of Drosha and Argonaute like subfamilies. According to research reports on miRNA pathways in P. falciparum, lacks the main candidates of Dicer complex and RISC in genome (21,22). Bioinformatics analysis of small RNA libraries could not identify Pf specific miRNAs in Pf-infected erythrocytes (21-23). Hence, to date there is not a single report on Plasmodium miRNAs.
Mobility of miRNAs within the original cell or to other cells, as well as within the own body is well known. As most of the miRNAs are conserved, in recent studies it has been observed that exchange/interactions of small RNA molecules/miRNAs that occur between the cells belong to different Kingdom. This phenomenon has been discovered during intracellular infection of pathogen, parasites, and symbionts (20,24). P. falciparum alters host cell physiology by remodulation of membrane proteomics. Expression of proteins such as Plasmodium falciparum 3D7 erythrocyte membrane protein 1 (PfEMP1), MSP family, Knob associated family and adhesive proteins occur at intervals to modify the host surface (25,26), which results in altered properties of the infected erythrocytes like cytoadherence, morphology and these molecules take over the overall charge of host cell machinery (26,27).
To find out the role of miR-451a, we targeted them on P. falciparum 3D7 transcriptome via in silico approach. This study revealed numerous interactions of miR-451a on crucial gene targets of P. falciparum, required for its growth, survival, and its pathogenesis.
Methods
Erythrocytic miRNAs found in Pf-infected erythrocytes (iEs) and its connection with the pathogenesis caused by P. falciparum were identified from the literature studies on PubMed and Scopus database. Among the various erythrocytic miRNAs, miR-451a focused (15,19-21,23,28-31) to study its regulatory role during host parasitic interactions.
Retrieval of data
Identified erythrocytic miRNA hsa-miR-451a-5p (Accession No.: MI0001729) was retrieved using miR Base-the miRNA database 22.1 (http://www.mirbase.org/). P. falciparum 3D7 transcriptome retrieved from PlasmoDB 8.0 (RefSeq assembly accession: GCF_000002765.5 latest).
Potential target prediction - cross kingdom approach
The selected miR-451a sequence was analysed against the P. falciparum transcriptome using psRNATarget tool (http://plantgrn.noble.org/psRNATarget/) (32) with default parameters including maximum expected threshold: 3, length of complementarity scoring: 20, maximum energy to unpair target site: 25 and translation inhibition will set between 9 and 11 nts, to identify the plausible human target genes. The Input method followed will be searching user-submitted small RNAs against the user-submitted transcripts.
Functional annotation and disease association of targeted Plasmodium Genes
Determination and prediction of the function(s) of miRNA-451a, which targeted the transcriptome of P. falciparum, gene ontology study carried out using g:Profiler (https://biit.cs.ut.ee/gprofiler) (33) software using the Gene Ontology (GO) terms. Functional enrichment analysis of the predicted Plasmodium target genes conducted in order to identify their role in molecular function (MF), biological process (BP) and cellular component (CC). To find protein class details encoded by P. falciparum targeted genes, PANTHER online tool (http://pantherdb.org/) (34) was performed. These parameters were statistically significant with the threshold P value <0.05.
Identification of protein-protein interactions and hub proteins
Protein-protein interactions were examined in The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org/) database (35), which is a well-known biological database and web resource to predict protein-protein interactions. Cytoscape software was also applied to visualize the protein interactions and analyse the hub proteins.
Pathway prediction analysis
In addition to gene ontology, pathways may be the important factors to investigate the role of the genes at cellular component synthesis, molecular functioning, biological processes and their downstream signalling. KEGG pathway enrichment analysis tool and literature review were used to identify the involvement of these targeted genes in various diseases/pathophysiological conditions.
Statistical analysis
It is not applicable in identification of erythrocytic miRNA, it’s retrieval, its potential target genes’ identification, protein-protein interactions of targeted genes and pathway prediction analysis.
Provisions of the Declaration of Helsinki (as revised in 2013)
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
On analysing the various datasets, it was observed that erythrocytic miRNAs predominantly interact with the Plasmodium transcriptome (20). During the infection, miRNAs translocate intracellularly from its cytoplasm to the parasitophorous vacuole (PV) and their enrichment in the P. falciparum infected erythrocytes shows strong correlation between them (20). In order to identify the possible interactions between miR-451a and P. falciparum mRNAs’ computational prediction software PSRNA tool was performed. Bioinformatics analysis revealed novel target genes through chimeric fusions of (hsa-miR-451a-5p: mRNAs of P. falciparum), which down-regulate the genes responsible for the invasion, pathogenesis, and survival of P. falciparum (Table 1).
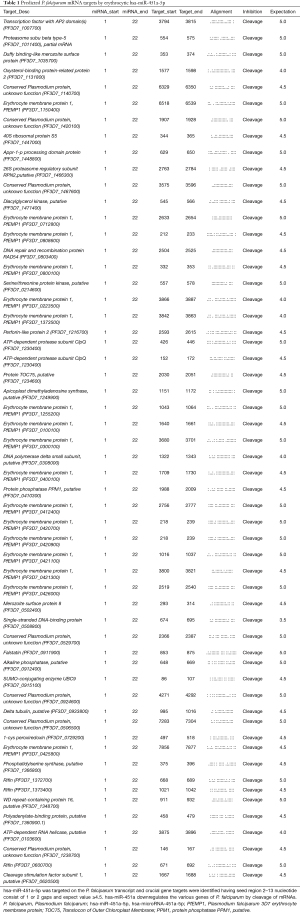
Full table
miRNA-target genes’ interactions
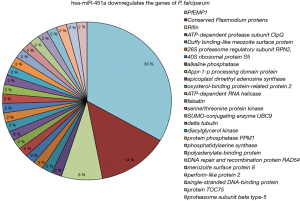
Considering the importance of miRNA-451a during host-parasitic interactions, it is essential to determine if any of the gene is having more than one targets for the mir-451a. As expected, number of targeting events appeared more than one in certain genes (Figure 1). These may be due to the length of the genes and having multiple complementary sites within the gene. Out of total targeted genes PfEMP1 gene contributed 33% participation (Figure 1). miR-451a showed multiple targets on the PfEMP1 gene and suppressed its expression by cleavage of PfEMP1 mRNA. After PfEMP1, uncharacterized genes with unknown functions were reported with maximum targets (Figure 1). These uncharacterized genes and their products add to the difficulties of recognizing the parasite biology. Rifin gene contributed to 8% with four targets within the gene (Figure 1). The remaining genes showed one target site for miR-451a (Figure 1).
Gene ontology of targeted P. falciparum genes
In order to characterize the crucial predicted gene products, GO categorization using g:Profiler Software (g:GOSt) (https://biit.cs.ut.ee/gprofiler/gost) was performed to investigate the MF, BP, CC and associated GO terms with hsa-miR-451a-5p targets (Figure 2).
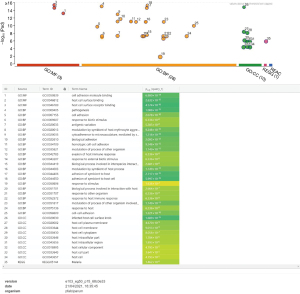
Figure 2 shows the annotation studies of targeted genes mainly contribute to molecular activities like cell adhesion molecule binding, host cell surface and its receptor binding. Whereas, biological processes include cell-cell adhesion, modulation of the host cell processes, response to stimuli/host’s immune response, antigenic variation, etc. Moreover, cellular components include intracellular components’ synthesis of the Plasmodium.
Further detailed analysis uncovered regarding the genes involved in each function mentioned below (Figure 3).

Functional studies determined the role of each targeted genes at the molecular, cellular and biological level. As per the list of genes mentioned in the MFs (Figure 3), it can be easily projected that PfEMP 1 having gene IDs: PF3D7_1150400, PF3D7_0223500, PF3D7_1255200) promotes all the three MFs of host cell surface receptor binding (GO ID: 0046789), host cell surface binding (GO ID: 0046812) and cell adhesion molecule binding (GO ID: 0050839) (Figure 3). These genes can efficiently rearrange the host cell membrane as well as developing strong intracellular network between PV and cytoplasm of erythrocytes to accomplish the erythrocytic life stages. Majority of miR-451a targeted genes code for the proteins which can efficiently promote BPs such as cell-cell adhesion (GO:0098609), cytoadherence (GO:0020035), antigenic variation (GO:0020033), erythrocytic aggregation (GO:0034117), sense and respond the external stimuli (GO:009605; GO:0009605; GO:0050896) including hosts’ immune response (GO:0052200; GO:0052572), etc. (Figure 3). miR-451a targeted genes mostly actively synthesize knob on the host cell surface (GO:0020030), cell membrane (GO:0020002), cytoplasmic content (GO:0030430; GO:0033643) and intracellular components (GO:0043656) as a part of cellular component (Figure 3).
Analysis of hsa-miR-451a targeted genes to obtain their protein class details using PANTHER classification system
The PANTHER database is designed for browsing ontology terms to retrieve associated protein families, subfamilies as well as individual protein. Targeted genes encoding proteins can be classified to a family or subfamily, and most of these are associated with significant MF or BP or CC classifications (Figure 4).
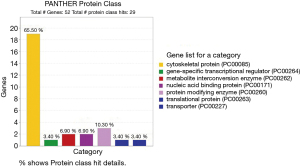
The total proteins class details retrieved from the targeted genes, cytoskeletal protein covers 65% (Figure 4), which is encoded by vital genes of P. falciparum such as PfEMP1 and Duffy binding-like (DBL) merozoite surface protein MSP8 (PF3D7_1035700). Targeted gene products except uncharacterized proteins have been classified under PANTHER protein class of PANTHER database and as mentioned in Table 2.
Full table
Proteins like tubulin, dorsal interacting protein 4, oxysterol-binding protein, ribosomal proteins and enzymes like ATP dependent RNA helicase, proteases are crucial targets to affect the growth and intracellular development of P. falciparum. Proteases and tubulin may participate in establishing intracellular network for the transport of nutrients and some essential bio molecules between the parasite and its host erythrocytes. These proteins can be key molecules to comprehend the host-parasitic information and help us to discover the targets of various anti-malarial bioactive compounds.
Protein-protein interactions and pathway prediction analysis of the miR-451a targeted genes
Comprehensive pathway prediction analysis was performed of the targeted genes of P. falciparum. Among the all genes, mainly genes were found principally associated with RNA degradation (map03018) and RNA transport (map03013) KEGG pathways. Whereas around 20 genes demonstrated a correlation with the KEGG:05144 Malaria pathway (https://www.kegg.jp/entry/pathway+map03018).
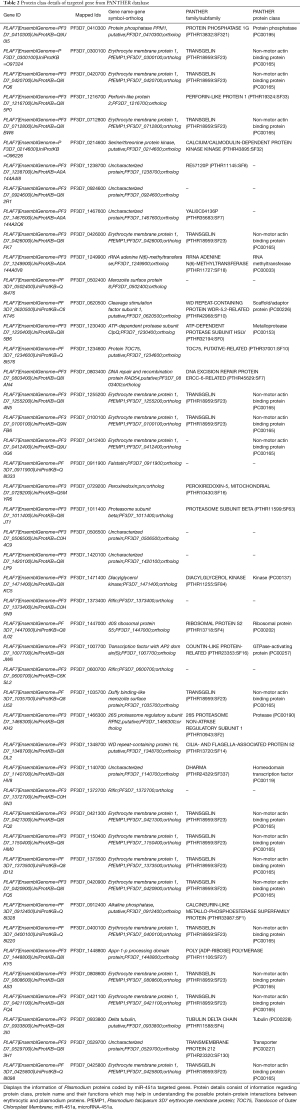
To identify the groups of interacting proteins and hub genes STRING database and Cytoscape software has been used. Out of total genes, 07 genes were found in protein-protein interactions. The input gene list has been mentioned below (Table 3) which involved in the protein-protein interactions (PPI) with the other genes and code for the protein domains belong to Helicase conserved superfamily C terminal domain region, Middle domain of eukaryotic initiation factor 4 G (eIFG4), epidermal growth factors, DEAD/DEAH box superfamily. While predicted functional protein partners in the PPI involved uncharacterized protein; DEAD box helicase 19 (PF14_0185), MSP 4, MSP 5 and uncharacterized protein; ATP-dependent RNA helicase, MIF4G domain containing protein—(PF11_0086), Aspargine rich protein—(MAL13P1.63), uncharacterized protein, ATP dependent RNA Helicase (PFL0100c), DnaJ/SEC63 protein (PF13_0102), RNA helicase (PF08_0111), uncharacterized protein eIF4, belong to the eukaryotic initiation factor 4E family (PFC0635c), uncharacterized protein DEAD/DEAH box Helicase (PF14_0370), merozoites surface protein 8—MSP8 (Figure 5).

Full table
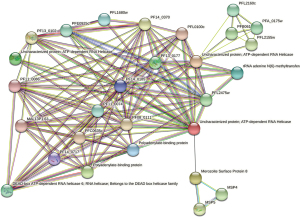
The hsa-miR-451a targeted crucial gene PfEMP1 showed PPI with the ten genes (Figure 6). Among these genes most of them are the variants of PfEMP1 (Figure 6). Rifin and gene belong to Var gene family are showed interactions and coexpression with PfEMP1.
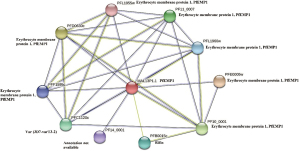
Protein domains description involve PFEMP DBL domain (PF03011), N-terminal segments of PfEMP1 (PF15447), acidic terminal segments, variant surface antigen of PfEMP1 (PF15445), Duffy binding domain (PF05424). These genes were associated with KEGG malaria pathway (map05144).
Discussion
To uncover the role of erythrocytic miRNA (miR-451a) mediated mRNA modulation in Plasmodium and to predict the cross-kingdom dynamics between different species, target identification and possible interactions between erythrocytic miRNA-Plasmodium mRNAs’ study carried out. Among the several erythrocytic miRNAs, miR-451 is extensively studied to understand the erythrocytic physiology under various pathophysiological conditions (19,20,23,29,36). Many studies have been conducted to determine its significance and to establish as a biomarker or therapeutic agents in various erythrocytes related disorders or diseased conditions including malaria (19,20,30,31,37). Translocation of ~100 erythrocytic miRNAs have been found in the PV formed by the parasite inside the erythrocytes. After parasite invasion, expression of miR-451 peaked after 32 hrs, indicating uptake of this miRNA during intra-erythrocytic cycle (38). It has been observed that miR-451 is located in the parasitophorous vacuole membrane (PVM) in infected erythrocytes (19,20), which suggests that translocation of these miRNAs to transcriptome of parasite takes place as a part of host protective mechanism. As per the miR-451a target prediction study, PfEMP1 and rifin genes contributed with multiple binding sites for miR-451a and their downregulation may affect the occurrence like cell-cell adhesion, antigenic variation, erythrocytic aggregation and host cell surface knob formation. Protein families associated with these genes are DBL adhesive domains, present on the surface of the iEs and mainly involve in erythrocyte invasion and cytoadherence of iEs. The genes (PfEMP1, rifin, MSP8, TOC75, oxysterol binding proteins, tubulin, DBL MSP) belonged to the formation and alteration of host’s intracellular, cytoplasmic and membrane components because of these protein details retrieved from the total targeted genes belong to the cytoskeletal proteins. Gene annotation and protein-protein interaction studies indicated mainly uncharacterized proteins. Beside the uncharacterized genes, chiefly genes (PF3D7_0620500, PF3D7_1007700, PF3D7_1447000, PF3D7_1448800, PF3D7_0915100, PF3D7_0103600, PF3D7_1360900.1, PF3D7_1249900) engaged in the RNA metabolism chiefly in RNA processing and degardation pathways. The active participation of these genes showed the importance of RNA metabolism due to the various intraerythrocytic life stages (trophozoites, schizonts, ring stages) specific protein expressions and its strict regulation. Erythrocytic miR-451a targeted P. falciparum genes that have a major role in governing various pathways include (I) vesicular trafficking pathway mediated by Perforin Like Protein 2, TOC75; (II) intracellular lipid/sterol pathway mediated by oxysterol binding protein which serve as a key molecule with varied functional relevance. More than one noteworthy genes belong to the key enzymes, those were found to be down regulated by miR-451 are (I) peroxidase (2-Cys peroxiredoxin); (II) phosphatases [alkaline phosphatase, protein phosphatase PPM1, putative (PPM1)]; (III) proteases ATP-dependent protease subunit ClpQ; (IV) proteases inhibitor (falstatin); (V) kinases (diacylglycerol kinase, serine threonine protein kinase); (VI) synthase (apicoplast dimethyl adenosine synthase); (VII) helicases (ATP-dependent RNA helicase); (VIII) polymerases (DNA polymerase delta small subunit). These key enzymes are thought to be essential regulators of numerous cells signaling pathways.
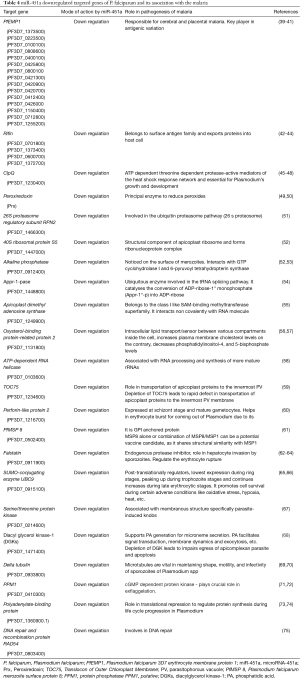
Detailed analysis of target genes and their products confirm their role in pathogenesis mentioned in Table 4.
Full table
Several reports indicated that, miR-451 is found to be elevated in sickle cell erythrocytes (19,76), and is associated with significantly reduced parasitaemia (19,20,77). miRNAs profile of HbAS and HbSS erythrocytes may significantly affect the parasite growth and survival (20,30,76,77). Thus, this may be the one of the protective mechanisms mediated by the host erythrocytes, which affects the intracellular parasite growth and survival (20). While the average levels of plasma miR-451 and miR-16 were significantly found lower in malaria patients (28). Moreover, transfection of miR-451 and miR-223 into P. falciparum iEs resulted into 50% reduction in parasitemia (20). This means miR-451a has strong relationship with the intracellular parasite growth and its survival (Figure 7) but, the expression of this miRNA can be crucial factor during the pathogensis and it will decide the fate of the diseased condition in the body.
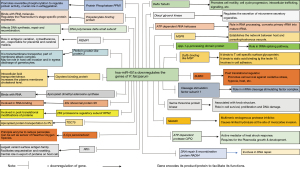
Now a days many bioinformatic analysis have been performed to understand the relationship between the dysregulation of miRNA and many infectious diseases (23,78,79). Based on this study it can be said that, erythrocytic miRNAs can modulate the gene expression via complementary binding on the target gene. This analysis predicts the significant outcome of a hsa-miR-451a (Figure 7). Based on this study, the mechanism behind altered miR 451a profile of sickle erythrocytes and their contribution in providing protection against plasmodial attack can also be easily understood miR-451.
Conclusions
Conclusively, the result indicated that miR-451a targeted maximally to the uncharacterized genes comprising unknown functions, which means no. of key protein molecules and its role in pathogenesis is yet to be discovered. Rest of the targeted genes and their products are involved mainly in the (I) modifications of erythrocyte membrane, cytoskeletal proteins to establish strong interactions with the host cell, as well as in the build-up of a strong intracellular network for the transportation of nutrients and release of waste materials; (II) RNA metabolism to regulate the synthesis of stage specific required proteins etc. Very few proteins are usually get involved in synthesis of transporters, their proliferation, response to external stimuli, DNA repair, etc. This shows the significance of miR-451a during the erythrocytic stages of P. falciparum. Enrichment of miR-451a in sickle cell anaemia can be one of the important initiatives of host defence mechanism to combat the parasite Plasmodium infection.
Additional in vitro as well as in vivo studies will be required to elucidate the mechanism of trans kingdom interactions related to erythrocytic miR-451a and P. falciparum transcript. Moreover, level of miR-451a inside the erythrocytes in normal as well as during various pathophysiological conditions and in the blood as form of extracellular vesicles (EVs) may provide key information about host-parasite complexity and the influence of its interaction with Plasmodium can be proved this molecule as a disease specific and accurate potential biomarker in malaria. Additionally, miRNA based novel therapeutic approach can also be designed in near future using the miRNA mimics or anti-miRs for the treatment of a variety of disease including malaria.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/exrna-21-5). All authors report laboratory and administrative assistance from Gujarat University. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization. World malaria report 2020. Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2020
- Sheriff O, Yaw A, Lai SK, et al. Plasmodium falciparum replication factor C subunit 1 is involved in genotoxic stress response. Cell Microbiol 2021;23:e13277 [Crossref] [PubMed]
- Mehra A, Balaji SN, Trivedi V. Hemozoin is a potential threat in cerebral malaria pathology through the induction of RBC-EC cytoadherence. Acta Trop 2021;217:105867 [Crossref] [PubMed]
- Amir A, Cheong FW, De Silva JR, et al. Diagnostic tools in childhood malaria. Parasit Vectors 2018;11:53. [Crossref] [PubMed]
- Reboud J, Xu G, Garrett A, et al. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc Natl Acad Sci U S A 2019;116:4834-42. [Crossref] [PubMed]
- Holmström O, Stenman S, Suutala A, et al. A novel deep learning-based point-of-care diagnostic method for detecting Plasmodium falciparum with fluorescence digital microscopy. PLoS One 2020;15:e0242355 [Crossref] [PubMed]
- Thu AM, Phyo AP, Landier J, et al. Combating multidrug-resistant Plasmodium falciparum malaria. FEBS J 2017;284:2569-78. [Crossref] [PubMed]
- Uwimana A, Legrand E, Stokes BH, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020;26:1602-8. [Crossref] [PubMed]
- Finotti A, Fabbri E, Lampronti I, et al. MicroRNAs and Long Non-coding RNAs in Genetic Diseases. Mol Diagn Ther 2019;23:155-71. [Crossref] [PubMed]
- Catanzaro G, Filardi T, Sabato C, et al. Tissue and circulating microRNAs as biomarkers of response to obesity treatment strategies. J Endocrinol Invest 2021;44:1159-74. [Crossref] [PubMed]
- Bobbin ML, Rossi JJ. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu Rev Pharmacol Toxicol 2016;56:103-22. [Crossref] [PubMed]
- Majumdar R, Rajasekaran K, Cary JW. RNA Interference (RNAi) as a Potential Tool for Control of Mycotoxin Contamination in Crop Plants: Concepts and Considerations. Front Plant Sci 2017;8:200. [Crossref] [PubMed]
- Babatunde KA, Yesodha Subramanian B, Ahouidi AD, et al. Role of Extracellular Vesicles in Cellular Cross Talk in Malaria. Front Immunol 2020;11:22. [Crossref] [PubMed]
- Mohammdai-asl J, Ramezani A, Norozi F, et al. MicroRNAs in erythropoiesis and red blood cell disorders. Frontiers in biology 2015;10:321-32. [Crossref]
- Sun L, Yu Y, Niu B, et al. Red Blood Cells as Potential Repositories of MicroRNAs in the Circulatory System. Front Genet 2020;11:442. [Crossref] [PubMed]
- Hamilton AJ. MicroRNA in erythrocytes. Biochem Soc Trans 2010;38:229-31. [Crossref] [PubMed]
- Byon JC, Papayannopoulou T. MicroRNAs: Allies or foes in erythropoiesis? J Cell Physiol 2012;227:7-13. [Crossref] [PubMed]
- Dore LC, Amigo JD, Dos Santos CO, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A 2008;105:3333-8. [Crossref] [PubMed]
- Chen SY, Wang Y, Telen MJ, et al. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One 2008;3:e2360 [Crossref] [PubMed]
- LaMonte G, Philip N, Reardon J, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012;12:187-99. [Crossref] [PubMed]
- Rathjen T, Nicol C, McConkey G, et al. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett 2006;580:5185-8. [Crossref] [PubMed]
- Xue X, Zhang Q, Huang Y, et al. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J 2008;7:47. [Crossref] [PubMed]
- Chamnanchanunt S, Fucharoen S, Umemura T. Circulating microRNAs in malaria infection: bench to bedside. Malar J 2017;16:334. [Crossref] [PubMed]
- Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet 2014;10:e1004602 [Crossref] [PubMed]
- de Koning-Ward TF, Dixon MW, Tilley L, et al. Plasmodium species: master renovators of their host cells. Nat Rev Microbiol 2016;14:494-507. [Crossref] [PubMed]
- Lavazec C. Molecular mechanisms of deformability of Plasmodium-infected erythrocytes. Curr Opin Microbiol 2017;40:138-44. [Crossref] [PubMed]
- Kilejian A. Interactions of Malaria Parasites and Their Host Erythrocytes. In: Moulder JW. editor. Intracellular Parasitism. Boca Raton: CRC Press 2020:213-25.
- Chamnanchanunt S, Kuroki C, Desakorn V, et al. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp Parasitol 2015;155:19-25. [Crossref] [PubMed]
- Wang T, Wu F, Yu D. miR-144/451 in hematopoiesis and beyond. ExRNA 2019;1:16. [Crossref]
- Verma HK, Ratre YK, Bhaskar LVKS, et al. Erythrocyte microRNAs: a tiny magic bullet with great potential for sickle cell disease therapy. Ann Hematol 2021;100:607-14. [Crossref] [PubMed]
- Zhang Y, Chu X, Wei Q. MiR-451 Promotes Cell Apoptosis and Inhibits Autophagy in Pediatric Acute Myeloid Leukemia by Targeting HMGB1. J Environ Pathol Toxicol Oncol 2021;40:45-53. [Crossref] [PubMed]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 2018;46:W49-54 [Crossref] [PubMed]
- Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191-8 [Crossref] [PubMed]
- Major JL, Bagchi RA, Pires da Silva J. Application of microRNA Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease. Methods Protoc 2020;4:5. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605-12. [Crossref] [PubMed]
- Wang W, Hang C, Zhang Y, et al. Dietary miR-451 protects erythroid cells from oxidative stress via increasing the activity of Foxo3 pathway. Oncotarget 2017;8:107109-24. [Crossref] [PubMed]
- Rangel G, Teerawattanapong N, Chamnanchanunt S, et al. Candidate microRNAs as Biomarkers in Malaria Infection: A Systematic Review. Curr Mol Med 2019;20:36-43. [Crossref] [PubMed]
- Garg S, Agarwal S, Kumar S, et al. Calcium-dependent permeabilization of erythrocytes by a perforin-like protein during egress of malaria parasites. Nat Commun 2013;4:1736. [Crossref] [PubMed]
- Jensen AR, Adams Y, Hviid L. Cerebral Plasmodium falciparum malaria: The role of PfEMP1 in its pathogenesis and immunity, and PfEMP1-based vaccines to prevent it. Immunol Rev 2020;293:230-52. [Crossref] [PubMed]
- McHugh E, Carmo OMS, Blanch A, et al. Role of Plasmodium falciparum Protein GEXP07 in Maurer’s Cleft Morphology, Knob Architecture, and P. falciparum EMP1 Trafficking. Mbio 2020;11:e03320-19. [Crossref] [PubMed]
- Obeng-Adjei N, Larremore DB, Turner L, et al. Longitudinal analysis of naturally acquired PfEMP1 CIDR domain variant antibodies identifies associations with malaria protection. JCI Insight 2020;5:137262 [Crossref] [PubMed]
- Mwakalinga SB, Wang CW, Bengtsson DC, et al. Expression of a type B RIFIN in Plasmodium falciparum merozoites and gametes. Malar J 2012;11:429. [Crossref] [PubMed]
- Goel S, Palmkvist M, Moll K, et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med 2015;21:314-7. [Crossref] [PubMed]
- Saito F, Hirayasu K, Satoh T, et al. Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors. Nature 2017;552:101-5. [Crossref] [PubMed]
- Ramasamy G, Gupta D, Mohmmed A, et al. Characterization and localization of Plasmodium falciparum homolog of prokaryotic ClpQ/HslV protease. Mol Biochem Parasitol 2007;152:139-48. [Crossref] [PubMed]
- Tschan S, Kreidenweiss A, Stierhof YD, et al. Mitochondrial localization of the threonine peptidase PfHslV, a ClpQ ortholog in Plasmodium falciparum. Int J Parasitol 2010;40:1517-23. [Crossref] [PubMed]
- Rathore S, Jain S, Sinha D, et al. Disruption of a mitochondrial protease machinery in Plasmodium falciparum is an intrinsic signal for parasite cell death. Cell Death Dis 2011;2:e231 [Crossref] [PubMed]
- Lilburn TG, Cai H, Gu J, et al. Exploring systems affected by the heat shock response in Plasmodium falciparum via protein association networks. Int J Comput Biol Drug Des 2014;7:369-83. [Crossref] [PubMed]
- Kawazu S, Komaki-Yasuda K, Oku H, et al. Peroxiredoxins in malaria parasites: parasitologic aspects. Parasitol Int 2008;57:1-7. [Crossref] [PubMed]
- Kimura R, Komaki-Yasuda K, Kawazu S, et al. 2-Cys peroxiredoxin of Plasmodium falciparum is involved in resistance to heat stress of the parasite. Parasitol Int 2013;62:137-43. [Crossref] [PubMed]
- Sridhar S, Bhat G, Guruprasad K. Analysis of bortezomib inhibitor docked within the catalytic subunits of the Plasmodium falciparum 20S proteasome. Springerplus 2013;2:566. [Crossref] [PubMed]
- Khosh-Naucke M, Becker J, Mesén-Ramírez P, et al. Identification of novel parasitophorous vacuole proteins in P. falciparum parasites using BioID. Int J Med Microbiol 2018;308:13-24. [Crossref] [PubMed]
- Wilkes JM, Doerig C. The protein-phosphatome of the human malaria parasite Plasmodium falciparum. BMC Genomics 2008;9:412. [Crossref] [PubMed]
- Kumaran D, Eswaramoorthy S, Studier FW, et al. Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme. Protein Sci 2005;14:719-26. [Crossref] [PubMed]
- Patron NJ, Waller RF, Archibald JM, et al. Complex protein targeting to dinoflagellate plastids. J Mol Biol 2005;348:1015-24. [Crossref] [PubMed]
- Zeng B, Zhu G. Two distinct oxysterol binding protein-related proteins in the parasitic protist Cryptosporidium parvum (Apicomplexa). Biochem Biophys Res Commun 2006;346:591-99. [Crossref] [PubMed]
- von Filseck J M, Mesmin B, Bigay J, et al. Building lipid ‘PIPelines’ throughout the cell by ORP/Osh proteins. Biochem Soc Trans 2014;42:1465-70. [Crossref] [PubMed]
- Chauhan M, Sourabh S, Yasmin R, et al. Biochemical characterization of Plasmodium falciparum parasite specific helicase 1 (PfPSH1). FEBS Open Bio 2019;9:1909-27. [Crossref] [PubMed]
- Sheiner L, Fellows JD, Ovciarikova J, et al. Toxoplasma gondii Toc75 Functions in Import of Stromal but not Peripheral Apicoplast Proteins. Traffic 2015;16:1254-69. [Crossref] [PubMed]
- Wirth CC, Glushakova S, Scheuermayer M, et al. Perforin-like protein PPLP2 permeabilizes the red blood cell membrane during egress of Plasmodium falciparum gametocytes. Cell Microbiol 2014;16:709-33. [Crossref] [PubMed]
- Alaro JR, Angov E, Lopez AM, et al. Evaluation of the immunogenicity and vaccine potential of recombinant Plasmodium falciparum merozoite surface protein 8. Infect Immun 2012;80:2473-84. [Crossref] [PubMed]
- Hadley T, Aikawa M, Miller LH. Plasmodium knowlesi: studies on invasion of rhesus erythrocytes by merozoites in the presence of protease inhibitors. Exp Parasitol 1983;55:306-11. [Crossref] [PubMed]
- Pandey KC, Singh N, Arastu-Kapur S, et al. Falstatin, a cysteine protease inhibitor of Plasmodium falciparum, facilitates erythrocyte invasion. PLoS Pathog 2006;2:e117 [Crossref] [PubMed]
- Sundararaj S, Saxena AK, Sharma R, et al. Cross-talk between malarial cysteine proteases and falstatin: the BC loop as a hot-spot target. PLoS One 2014;9:e93008 [Crossref] [PubMed]
- Issar N, Roux E, Mattei D, et al. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol 2008;10:1999-2011. [Crossref] [PubMed]
- Reiter K, Mukhopadhyay D, Zhang H, et al. Identification of biochemically distinct properties of the small ubiquitin-related modifier (SUMO) conjugation pathway in Plasmodium falciparum. J Biol Chem 2013;288:27724-36. [Crossref] [PubMed]
- Kun JF, Hibbs AR, Saul A, et al. A putative Plasmodium falciparum exported serine/threonine protein kinase. Mol Biochem Parasitol 1997;85:41-51. [Crossref] [PubMed]
- Bullen HE, Jia Y, Yamaryo-Botté Y, et al. Phosphatidic Acid-Mediated Signaling Regulates Microneme Secretion in Toxoplasma. Cell Host Microbe 2016;19:349-60. [Crossref] [PubMed]
- Sinden RE, Talman A, Marques SR, et al. The flagellum in malarial parasites. Curr Opin Microbiol 2010;13:491-500. [Crossref] [PubMed]
- Spreng B, Fleckenstein H, Kübler P, et al. Microtubule number and length determine cellular shape and function in Plasmodium. EMBO J 2019;38:e100984 [Crossref] [PubMed]
- Guttery DS, Poulin B, Ramaprasad A, et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell Host Microbe 2014;16:128-40. [Crossref] [PubMed]
- Bennink S, Kiesow MJ, Pradel G. The development of malaria parasites in the mosquito midgut. Cell Microbiol 2016;18:905-18. [Crossref] [PubMed]
- Minns AM, Hart KJ, Subramanian S, et al. Nuclear, Cytosolic, and Surface-Localized Poly(A)-Binding Proteins of Plasmodium yoelii. mSphere 2018; [Crossref] [PubMed]
- Bennink S, Pradel G. The molecular machinery of translational control in malaria parasites. Mol Microbiol 2019;112:1658-73. [Crossref] [PubMed]
- Gopalakrishnan AM, Kumar N. Opposing roles for two molecular forms of replication protein A in Rad51-Rad54-mediated DNA recombination in Plasmodium falciparum. mBio 2013;4:e00252-13. [Crossref] [PubMed]
- Duraisingh MT, Lodish HF. Sickle cell microRNAs inhibit the malaria parasite. Cell Host Microbe 2012;12:127-8. [Crossref] [PubMed]
- Bunn HF. The triumph of good over evil: protection by the sickle gene against malaria. Blood 2013;121:20-5. [Crossref] [PubMed]
- Kooshkaki O, Rezaei Z, Rahmati M, et al. MiR-144: A New Possible Therapeutic Target and Diagnostic/Prognostic Tool in Cancers. Int J Mol Sci 2020;21:2578. [Crossref] [PubMed]
- Wang F, Ling L, Yu D. MicroRNAs in β-thalassemia. The American Journal of the Medical Sciences 2021;362:5-12. [Crossref] [PubMed]
Cite this article as: Joshi U, Patel M, Pandya H, George LB, Highland H. Functional prediction of human erythrocytic miR-451a on Plasmodium falciparum 3D7 transcriptome—an in-silico study. ExRNA 2021;3:3.


