
Novel easy-to-use approach to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-carrying extracellular vesicles in plasma
Introduction
The difficult times of the coronavirus disease 2019 (COVID-19) pandemic have necessitated the rapid development of sensitive diagnostic tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. According to clinical practice guided by World Health Organization (WHO), Centers for Disease Control and Prevention (CDC), European Centre for Disease Prevention and Control (ECDC), and related global health institutions, the selection of SARS-CoV-2 test should be based on the wider context including both the prevalence of the virus variants in the population and the status of the tested individual.
Laboratory diagnostic testing for SARS-CoV-2 infection—difficulties and possible improvements
Institutionally authorized specific diagnostic tests comprise those detecting either viral RNA (nucleic acid amplification tests, NAATs) and antigens (immunoassays) or specific antibodies (serological testing) in different materials. While the serological assays have a very limited indications for diagnostic testing of current infection, the recommended, and thus the most widely used NAAT, i.e., reverse transcription quantitative PCR (RT-PCR), allows for highly sensitive detection of viral RNA in respiratory specimens (1).
It is worth noting that SARS-CoV-2 is one of the few human respiratory viruses that replicates in both upper and lower respiratory tracts (2). Consequently, virus shedding dynamics differs across the respiratory tract, in most cases remaining higher and prolonged in lower respiratory tract, while declining fastly in the upper parts after infection (3). Thus, there is a quite high risk of false negative results while testing nasopharyngeal swabs, the most commonly collected material (4). On the other hand, lower respiratory tract specimens are more difficult to obtain on an outpatient basis, but appear critical for proper diagnosis of severely ill patients especially (2). In addition, detection of viral RNA in plasma begins to be considered in COVID-19 diagnosis and patient monitoring (5), especially that SARS-CoV-2 RNAemia seems to correlate strongly with disease severity and clinical outcome (6,7). However, some studies showed the very low positive rate of blood samples (4). One can speculate that this may be due to a high plasma dilution of viral RNA.
Most NAATs are preceded by an RNA extraction. This step extends the execution time and requires the specific reagents, which global shortages happen throughout the pandemic (8). Therefore, at present, attempts are undertaken to shorten and simplify NAATs by removing the RNA extraction step (8). Some reports suggest that this could be achieved by simple direct heating of specimens before amplifying viral RNA genes with NAAT (9-11). But this approach is still fraught with many challenges, including optimalization across specimen types (8). For instance, one can speculate that RNA dilution in plasma may make heating insufficient to obtain enough nucleic acids for further amplification.
Along these lines, a different and very promising approach has been recently proposed by Ning et al. (12). The authors based their idea on the fact that a part of the plasma pool of SARS-CoV-2 nucleic acids is transmitted by extracellular vesicles (EVs). As discussed below, such a transfer is suggested to facilitate viral spreading and immune evasion, which aggravates the disease course and may cause the extrapulmonary complications (13). On the other hand, isolation of RNA-loaded EVs from diagnostic material is supposed to increase the amount and purity of yielded nucleic acids for further analysis.
Novel approach to detect viral RNA in plasma EVs
Based on the latter, the authors (12) developed a novel assay combining two already established technologies, i.e., EV’s isolation through binding by immobilized specific antibodies as well as liposome-mediated reagent delivery. At the first step, EVs are directly captured from plasma samples with the use of specific antibodies against their surface marker CD81 that had been coated onto the enzyme-linked immunosorbent assay (ELISA) plate wells. Then, liposomes that contain all reagents essential for (I) reverse transcriptase (RT); (II) recombinase polymerase amplification (RPA); and (III) clustered regularly interspaced short palindromic repeat (CRISPR)-Cas12a reactions (8), are added to the plate wells to fuse with captured EVs. Finally, the resulting CRISPR-FDS fluorescent signal is easily measured with a benchtop plate reader at 480 nm excitation and 530 nm emission wavelengths (Figure 1). The fusion process was controlled with transmission electron microscopy, and confirmed in a Fӧrster resonant energy transfer (FRET) dequenching assay. It should be stressed that this assay does not require the extraction of RNA, in contrast to another newly developed strategy so-called extracellular vesicle capture by antibody of choice and enzymatic release (EV‐CATCHER) allowing for subsequent sequencing of small RNA molecules (14).
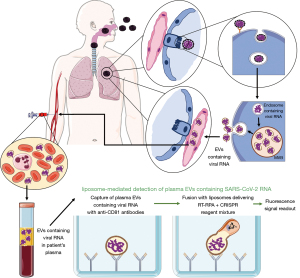
Afterwards, the authors were able to successfully distinguish the patients with COVID-19 from those with a negative RT-qPCR result of nasopharyngeal swab testing with the use of newly developed assay, and then proved its specificity by assaying the samples containing RNA from other human respiratory viruses. Interestingly, this assay allowed to detect EV-contained SARS-CoV-2 RNA in archived plasma obtained from six hospitalized patients with pulmonary lesions consistent with COVID-19 that had the negative RT-qPCR results. Moreover, it confirmed the long-lasting presence of SARS-CoV-2 RNA in plasma EVs obtained from two young children that had negative nasal swab results at all time points but positive results for specific IgGs. Finally, in a non-human primate model of SARS-CoV-2 infection, Ning et al. (12) showed that viral RNA expression time course in plasma EVs exhibited a delayed and sustained peak, when compared to results from nasal swabs testing. Altogether, these findings suggest that the newly developed assay allows to detect SARS-CoV-2 RNA in plasma EVs with a high sensitivity not only early after infection, but also when the results of nasopharyngeal swabs testing are or become again negative. Thus, after successful optimalization, this assay becomes a promising candidate for the general use at clinical conditions. However, some aspects have to be addressed in detailed to increase the clinical significance of the assay.
The origin of viral RNA detected in plasma EVs—should it be handled with care?
As pointed out by the authors (12), the form of the assay-detected EV RNA cargo remains unclear. At present, it is difficult to determine whether the detected RNA derived either from the whole or the part of viral genome, or from the whole virion. The latter, however, seems to be less possible since reaction conditions were unlikely to promote the release of virion-encapsulated RNA as well as the initial studies failed to detect viral proteins in proteomic analysis of plasma EVs (12). Therefore, one can speculate that the isolated EVs contain RNA material, but not the intact virion of the new coronavirus. Consequently, this provokes other important questions for consideration, i.e., if EVs loaded with SARS-CoV-2 RNA are infectious and how the nucleic acid cargo was packaged into EVs.
Along these lines, it has already been demonstrated that viruses may modulate both the EV’s biogenesis in infected cell and the vesicle content (15). As a result, EVs released by infected cells may contain virus-derived components that could either drive or limit viral replication in targeted cells. Moreover, growing evidence indicates that both enveloped and non-enveloped viruses may exploit the endocytic pathway that is physiologically used for EV’s biogenesis to produce virions that, after secretion, are capable of infecting the targeted cells (16). Thus, it has been assumed that transmission by EVs may not only facilitate virus invasion of entry-receptor-expressing cells but also of bystander cells that normally would avoid infection, thereby passing the infection to secondary tissues. In addition, EV-transmitted factors were suggested to promote sepsis and multi-organ failure in critically ill COVID-19 patients (15,17). Finally, incorporation into EVs may also prevent the viral particle from binding by neutralizing antibodies and further degradation by immune cells (16). Altogether, these findings strongly imply the deleterious role of EVs in infection spreading and immune evasion by viruses, which in turn may aggravate the course of the disease and its complications. Thus, one can assume that EVs loaded with whole viral genome are infectious in vivo, and recent data implies that they may preserve the infectivity in vitro (18), likely also after isolation from biological material. Hence, these aspects require further investigation as they have a critical importance for biosafety in laboratories and for public health. In addition, it is worth noting that some of the methods standardly used for EV’s isolation have already been reported to co-isolate viral particles (16,19), making EVs highly contagious. These findings justify the recommendation to use the heightened laboratory safety measures while isolating EVs from specimens collected from COVID-19 patients (16).
On the other hand, EV’s enrichment by CD63 bead affinity purification was shown to eliminate the viral particles from yielded CD63-positive vesicle fraction (19). This suggests that in the discussed study (12), EV’s fraction captured by anti-CD81 antibodies do not contain co-isolated viral particles. Additionally, this assumption is in line with the abovementioned observation that CD81-enriched EVs did not contain viral proteins (12). Thus, it could be assumed that the detected viral RNA was indeed carried by isolated EVs, and not artificially co-isolated during vesicle separation. However, there is still an open question if the detected RNA comes from the whole viral genome or from its fragments.
Interpreting the results
Similarly to NAATs, the results of currently developed assay are interpreted qualitatively and considered positive when the measured fluorescence signal is equal or greater than the mean signal of the negative controls plus three times their standard deviation (12). The attempts should be taken to allow the quantitative interpretation of the data, since one can speculate that the number of coronavirus RNA-positive EVs in plasma varies across the infection course and may possibly correlate with some clinical measures, such as the disease severity and the risk of extrapulmonary complications.
Furthermore, the currently developed assay is designed to isolate the total pool of CD81-positive EVs from plasma without discriminating them according to other parameters, such as the cellular and tissue sources and physicochemical properties. Although this approach appears enough for diagnostic purposes, several aspects have to be taken into consideration; i.e., (I) the yielded EV’s fraction is very heterogeneous; (II) the percentage of captured EVs that contain viral RNA remains impossible to estimate; and (III) the viral RNA load levels can vary among EV subtypes. Importantly, this diversity might impact the clinical significance of the assay. Therefore, further investigation is needed to determine if the obtained results correspond to clinical status of infection as well as how to co-interpret them with patient condition. Accordingly, the authors (12) reported the prolonged detectability of SARS-CoV-2 RNA in plasma EVs in comparison to swab samples, making it a promising candidate for a more durable marker of infection. However, one can speculate that the positive result may indicate not only the current active infection when the patient is highly contagious, but also the post-infection stage when cells may possibly remove waste viral RNA by packaging it into EVs, and even the late/latent phase after integration of viral genetic material into host cell genome. The authors (12) seem to be aware of this possibility as they have undertaken the studies improving their assay to enable the detection of the subgenomic RNA fragments that may serve as an indicator of an active infection. This should greatly support the attempts to distinguish these infection phases from one another.
Besides, an alternate pathway for a transfer of extracellular RNA between cells was formerly shown, and involved the association of freely circulating miRNA with EVs outside the cell (20). Taking this possibility into account for viral RNA, it could be hypothesized that free viral RNA may associate with plasma EVs in circulation during RNAemia. Such an EV-associated RNA pool could also be then detected with the currently discussed liposome-based assay (12). However, this hypothesis requires further investigation.
Biological significance of EV-loaded viral RNA
From another point of view, the therapeutic effect of convalescent plasma was proposed to be facilitated by the contained EVs (21). However, stable detection of SARS-CoV-2 RNA in plasma EVs during the long time course, as shown even up to 60 days from the initial evaluation (12), strongly suggests that EVs containing nucleic acids of the new coronavirus are present in convalescent plasma, and thus could impact its therapeutic potential. Accordingly, one can speculate that such EV-enclosed viral RNA fragments may mimic adjuvant activity and thus increase the anti-viral immune responses. On the other hand, EVs carrying multiple whole viral genomes may constitute an infectious unit (22) that might possibly pass the infection to convalescent plasma recipient thereby worsening the clinical outcome. The discussed assay allowing to detect viral RNA transmitted by EVs appears profoundly useful to investigate these hypotheses, especially when improved to distinguish the whole viral genome from its likely noninfectious fragments.
Concluding the advantages and disadvantages of clinical application of discussed assay
The main advantages of newly developed assay include its simplicity, ease of obtaining diagnostic material, quite short duration, lack of RNA extraction step, commonly used result reading method. However, before its widespread use, some disadvantages need to be improved. The reagent-loaded liposome supply chain could be developed to increase assay reproducibility. The form of viral RNA in EVs should be recognized to judge whether it is infectious or not. Attempts should be undertaken to detect SARS-CoV-2 RNA in EVs isolated from other materials, such as bronchoalveolar lavage fluid and exhaled breath condensate. Future studies are also required to assess the putative correlation of assay results with patient condition, infection course and severity, and with the risk of complications. The latter would greatly increase the clinical significance of the discussed approach.
Considering the improvement of the above discussed limitations of the commented assay, it may provide a promising complementary tool supporting the rapid diagnostic procedure, especially of symptomatic patients with negative swab results, asymptomatic individuals with inconclusive RT-PCR results as well as for immunocompromised patients. Moreover, it may potentially be implemented to point-of-care testing, which increases its clinical significance. Finally, when improved, the assay may serve as an important extraction-free approach for tracking the infectiveness of EV-contained viral RNA in various biological samples.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/exrna-21-15). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- da Silva SJR, Silva CTAD, Guarines KM, et al. Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19. ACS Infect Dis 2020;6:2319-36. [Crossref] [PubMed]
- Vu MN, Menachery VD. Binding and entering: COVID finds a new home. PLoS Pathog 2021;17:e1009857 [Crossref] [PubMed]
- Alidjinou EK, Poissy J, Ouafi M, et al. Spatial and Temporal Virus Load Dynamics of SARS-CoV-2: A Single-Center Cohort Study. Diagnostics (Basel) 2021;11:427. [Crossref] [PubMed]
- Bwire GM, Majigo MV, Njiro BJ, et al. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol 2021;93:719-25. [Crossref] [PubMed]
- Azghandi M, Kerachian MA. Detection of novel coronavirus (SARS-CoV-2) RNA in peripheral blood specimens. J Transl Med 2020;18:412. [Crossref] [PubMed]
- Jacobs JL, Bain W, Naqvi A, et al. SARS-CoV-2 Viremia is Associated with COVID-19 Severity and Predicts Clinical Outcomes. Clin Infect Dis 2021; Epub ahead of print. [Crossref] [PubMed]
- Rodríguez-Serrano DA, Roy-Vallejo E, Zurita Cruz ND, et al. Detection of SARS-CoV-2 RNA in serum is associated with increased mortality risk in hospitalized COVID-19 patients. Sci Rep 2021;11:13134. [Crossref] [PubMed]
- Mardian Y, Kosasih H, Karyana M, et al. Review of Current COVID-19 Diagnostics and Opportunities for Further Development. Front Med (Lausanne) 2021;8:615099 [Crossref] [PubMed]
- Alcoba-Florez J, González-Montelongo R, Íñigo-Campos A, et al. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int J Infect Dis 2020;97:66-8. [Crossref] [PubMed]
- Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill 2020;25:2000398 [Crossref] [PubMed]
- Lübke N, Senff T, Scherger S, et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol 2020;130:104579 [Crossref] [PubMed]
- Ning B, Huang Z, Youngquist BM, et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat Nanotechnol 2021;16:1039-44. [Crossref] [PubMed]
- Xia X, Yuan P, Liu Y, et al. Emerging roles of extracellular vesicles in COVID-19, a double-edged sword? Immunology 2021;163:416-30. [Crossref] [PubMed]
- Mitchell MI, Ben-Dov IZ, Liu C, et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J Extracell Vesicles 2021;10:e12110 [Crossref] [PubMed]
- Gurunathan S, Kang MH, Kim JH. Diverse Effects of Exosomes on COVID-19: A Perspective of Progress From Transmission to Therapeutic Developments. Front Immunol 2021;12:716407 [Crossref] [PubMed]
- Nunez Lopez YO, Casu A, Pratley RE. Investigation of Extracellular Vesicles From SARS-CoV-2 Infected Specimens: A Safety Perspective. Front Immunol 2021;12:617042 [Crossref] [PubMed]
- Clauss M, Chelvanambi S, Cook C, et al. Viral Bad News Sent by EVAIL. Viruses 2021;13:1168. [Crossref] [PubMed]
- Kerviel A, Zhang M, Altan-Bonnet N. A New Infectious Unit: Extracellular Vesicles Carrying Virus Populations. Annu Rev Cell Dev Biol 2021;37:171-97. [Crossref] [PubMed]
- Chugh PE, Sin SH, Ozgur S, et al. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog 2013;9:e1003484 [Crossref] [PubMed]
- Bryniarski K, Ptak W, Martin E, et al. Free Extracellular miRNA Functionally Targets Cells by Transfecting Exosomes from Their Companion Cells. PLoS One 2015;10:e0122991 [Crossref] [PubMed]
- Askenase PW. COVID-19 therapy with mesenchymal stromal cells (MSC) and convalescent plasma must consider exosome involvement: Do the exosomes in convalescent plasma antagonize the weak immune antibodies? J Extracell Vesicles 2020;10:e12004 [Crossref] [PubMed]
- Leeks A, Sanjuán R, West SA. The evolution of collective infectious units in viruses. Virus Res 2019;265:94-101. [Crossref] [PubMed]
Cite this article as: Nazimek K. Novel easy-to-use approach to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-carrying extracellular vesicles in plasma. ExRNA 2021;3:5.


