
A novel circular RNA acts as a key modulator of epidermal growth factor receptor signaling in non-small cell lung cancer
One attractive chemotherapeutic target in efforts to defeat non-small cell lung cancer (NSCLC) is the epidermal growth factor receptor (EGFR) and its associated signaling mechanisms. The EGFR is a transmembrane tyrosine kinase receptor that is typically abnormally activated during NSCLC and which signals through a variety of canonical signaling pathways including RAS/RAF/MEK/ERK, JAK/STAT, and PI3K/Akt/mTOR/p70S6K (1). The many molecular targets integrated into EGFR signaling have led to the development of a variety of chemotherapeutic agents that can prevent its oncogenic function including blockers and tyrosine kinase inhibitors of the EGFR, and inhibitors of its downstream kinase effectors (2-5). Despite these pharmacologic efforts, lung cancer remains the world’s leading cause of cancer deaths (6) and due to mutations in EGFR pathway proteins, chemotherapeutic resistance frequently occurs in NSCLC making prognoses unfavorable (7).
The difficulties encountered with designing chemotherapeutic strategies against NSCLC signaling proteins that often undergo mutation might be resolved if more stable non-protein drug targets exist in the EGFR pathway. This idea has led to an emerging research focus on the role of noncoding RNAs in lung cancer. Recently, dysregulation of noncoding circular RNAs (circRNAs) has been shown to be implicated in many cancers including lung cancer (8,9). CircRNAs are closed loop RNAs produced by RNA binding proteins that mediate the backsplicing of pre-mRNA transcripts; these circRNAs can then be subsequently degraded by RNase L (10-12). CircRNAs represent a form of gene silencing as they are able to bind to and suppress microRNAs (miRNAs) that in turn bind to mRNA transcripts preventing their translation into proteins (13). Suppression of pathologically upregulated circRNAs can be accomplished using clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 13 (Cas13) methods (14). Cas13 proteins are ribonucleases which form complexes with short CRISPR RNAs (crRNA) that have sequences complementary to targeted single-stranded RNA transcripts which are then degraded by Cas13’s nuclease activity (14,15). The CRISPR/Cas13 system has little potential for causing off-target effects allowing it to act as a high-precision therapeutic unlike more traditional forms of RNA interference mediated gene silencing (14). An additional advantage of CRISPR/Cas is the ability to use it in both cell-based and animal models (16).
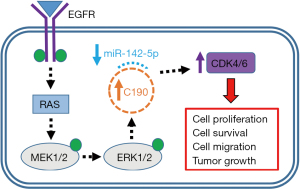
In the study of Ishola et al. (17), the authors analyzed the mechanistic function in NSCLC of the circular noncoding RNA, hsa_circ_0000190 (C190), a circRNA previously identified as a negative prognostic biomarker of lung cancer (18). They first showed that C190 is upregulated in clinical lung cancer tissues and cancer cell lines; whereas, the expression of CNIH4, the linear product of the C190 gene, was unaltered. Then, they found that activation of EGFR signaling caused altered C190 expression primarily through the MAPK/ERK and not JAK/STAT or PI3K/Akt/mTOR/p70S6K pathways. Further, overexpressing C190 promoted tumor cell growth and migration in vitro and in vivo and activated several cyclin dependent kinases (CDKs) linked with cancer progression (Figure 1). Next, the authors utilized a CRISPR/Cas13a system where NSCLC cells stably expressing Cas13a were transfected with either control or C190 targeting crRNAs and then xenografted into mice to demonstrate that C190 knockdown decreased tumor growth in vivo. As a final set of experiments, they showed that C190 likely targets CDKs by suppressing the microRNA, miR-142-5p. An important point suggested by the authors is that their CRISPR/Cas13a technique could be used to precisely suppress C190 in NSCLC without causing off-target effects and would still function against NSCLC independently of the effects of protein mutations that frequently occur in the EGFR pathway components of this disease.
There are several issues raised in the study of Ishola et al. that require further consideration. NSCLC represents a heterogeneous group of cancers that can integrate non-EGF receptor mediated genes, e.g., K-RAS and EML4-ALK which can incorporate signaling mechanisms outside of the MAPK/ERK, JAK/STAT or PI3K/Akt/mTOR/p70S6K pathways considered by the authors (19,20). Therefore, additional research would seem necessary to determine if C190 regulation is truly central in the treatment of all forms of NSCLC. Also, the study’s analysis of C190 signaling is primarily derived from cell-based methods which follow short-term time scales that may not identify later compensatory signaling behaviors which could modulate or counteract C190 and its related mechanisms. Interestingly, in prior work (18), the authors suggested that C190 targets miR-767-5p, miR-382-5p, miR-382-3p, and miR-1299, other than the miR-142-5p analyzed in the current study. This data suggests that C190 may act within a larger microRNA network to modulate pleiotropic functions with uncertain signaling effects. Additional research is requisite to determine how suppression of C190 levels would affect wider microRNA systems biology in NSCLC. Further, as C190 is also upregulated in other cancers, e.g., gastric cancer (21), expression of this circRNA may be fairly ubiquitous in human tissues which could mean that its suppression via CRISPR/Cas13a based methods might cause deleterious consequences in some normal non-lung tissues. Finally, as a translational method, a precision NSCLC targeted form of the CRISPR/Cas technique proposed in the study of Ishola et al. would seem to be incumbent to prevent off-target effects in non-NSCLC tissues where other microRNAs may be regulated by C190.
AcknowledgmentsOther Section
Funding: None.
FootnoteOther Section
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-7/coif). YG is supported by a National Institute of Health grant (P20 GM104357), by a COBRE/MS CEPR National Institute of Health grant (P20GM121334) and by a National Institute of Health grant (R01DE029803). YG and JDM are supported by a Cancer Center and Research Institute pilot grant, and all funding under this grant is dispensed to the University of Mississippi Medical Center and not to the author of this manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Farnsworth DA, Chen YT, de Rappard Yuswack G, et al. Emerging Molecular Dependencies of Mutant EGFR-Driven Non-Small Cell Lung Cancer. Cells 2021;10:3553. [Crossref] [PubMed]
- Cheng H, Shcherba M, Pendurti G, et al. Targeting the PI3K/AKT/mTOR pathway: potential for lung cancer treatment. Lung Cancer Manag 2014;3:67-75. [Crossref] [PubMed]
- Tricker EM, Xu C, Uddin S, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov 2015;5:960-71. [Crossref] [PubMed]
- Mohrherr J, Haber M, Breitenecker K, et al. JAK-STAT inhibition impairs K-RAS-driven lung adenocarcinoma progression. Int J Cancer 2019;145:3376-88. [Crossref] [PubMed]
- Amelia T, Kartasasmita RE, Ohwada T, et al. Structural Insight and Development of EGFR Tyrosine Kinase Inhibitors. Molecules 2022;27:819. [Crossref] [PubMed]
- World Health Organization. International Agency for Research on Cancer. Cancer Today. 2020. Available online: https://gco.iarc.fr/today/online-analysis
- Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B 2015;5:390-401. [Crossref] [PubMed]
- Qiu M, Xia W, Chen R, et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res 2018;78:2839-51. [Crossref] [PubMed]
- Chen T, Luo J, Gu Y, et al. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer 2019;10:930-41. [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015;29:2168-82. [Crossref] [PubMed]
- Liu CX, Li X, Nan F, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019;177:865-880.e21. [Crossref] [PubMed]
- Liu Y, Ao X, Yu W, et al. Biogenesis, functions, and clinical implications of circular RNAs in non-small cell lung cancer. Mol Ther Nucleic Acids 2022;27:50-72. [Crossref] [PubMed]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature 2017;550:280-4. [Crossref] [PubMed]
- East-Seletsky A, O'Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016;538:270-3. [Crossref] [PubMed]
- Gao J, Luo T, Wang J. Gene interfered-ferroptosis therapy for cancers. Nat Commun 2021;12:5311. [Crossref] [PubMed]
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res 2022;82:75-89. [Crossref] [PubMed]
- Luo YH, Yang YP, Chien CS, et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers (Basel) 2020;12:1740. [Crossref] [PubMed]
- Bayliss R, Choi J, Fennell DA, et al. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci 2016;73:1209-24. [Crossref] [PubMed]
- Xie M, Xu X, Fan Y. KRAS-Mutant Non-Small Cell Lung Cancer: An Emerging Promisingly Treatable Subgroup. Front Oncol 2021;11:672612. [Crossref] [PubMed]
- Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 2017;466:167-71. [Crossref] [PubMed]
Cite this article as: Monroe JD, Gibert Y. A novel circular RNA acts as a key modulator of epidermal growth factor receptor signaling in non-small cell lung cancer. ExRNA 2022;4:11.


