
The extracellular miRNA fingerprint of kidney disease: a narrative review
Introduction
MicroRNA (miRNA) biogenesis and function
Noncoding RNAs (ncRNAs) have arisen as a new paradigm in gene regulation and cell differentiation (1). Further of ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) involved in protein synthesis, regulatory ncRNAs classify based on their size: those shorter than 200 nucleotides (nt) are called short non-coding RNAs and include miRNAs, short interfering RNAs (siRNAs), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA) and piwi-interacting RNAs (piRNAs), whereas those longer than 200 nt are called long non-coding RNAs (lncRNAs) and include linear [long intergenic (lincRNA), intronic RNA, enhancer RNA (eRNA), natural antisense transcripts (NAT)] and circular RNA (circRNA) (2). They can regulate gene expression at transcriptional or post-transcriptional level by modulating chromatin structure, RNA maturation and protein synthesis/transport (3).
MiRNAs are a large family of conserved, small, non-coding RNAs of 19–25 nt long that repress the translation and/or degradation of their target mRNAs (4). Since the discovery of the first miRNA, Lin-4, in Caenorhabditis elegans in 1993, thousands of miRNAs have been uncovered in many multicellular organisms (5,6). At present, there are more than 2,600 and 1,900 mature miRNAs described in human and mouse, respectively (7). This regulatory system is based on base-pair complementarity between miRNAs and target sequences mainly located in the 3' untranslated region (UTR) of mRNAs. However, functional miRNA binding sites can also be found in the 5' UTR and open reading frame regions (ORFs) (8). This feature makes possible that a single miRNA can potentially target hundreds mRNAs and that one mRNA can be regulated by several miRNAs, with cooperative repression achieved by binding closely spaced target sites (9). This dynamic interaction relies on many factors, including the subcellular location, the abundance of miRNAs and target mRNAs and the affinity of miRNA-mRNA interactions (10). MiRNAs are transcribed by RNA polymerase II as a long precursor RNA primary miRNA (pri-miRNA) which is processed by the DROSHA-DGCR8 in the nucleus, resulting in a precursor miRNA (pre-miRNA) of ~70 nt in length. Once exported into the cytoplasm, it is shortened by DICER, yielding a ~22-nt mature miRNA. There, miRNAs associate with specific mRNAs within the multiprotein complex of Argonaute proteins, the core of the RNA-induced silencing complex (RISC) (11). One strand of the miRNA (‘guide strand’) is loaded into argonaute (AGO), whereas the other strand (‘passenger strand’) is eliminated. Alternative cleavage by DROSHA or DICER leads to the generation of isomiRs (9). Further, multiple non-canonical miRNA biogenesis pathways have been discovered, which include Drosha/DGCR8- and Dicer-independent processing routes (10). MiRNAs show very specific expression patterns that differ among tissues and cell types and are involved in virtually every cellular process, including development, differentiation, stress response and apoptosis (12).
Extracellular miRNAs
Although miRNAs function cell-intrinsically, miRNAs can be found in mostly every body fluid which support their role in the communication between cells and tissues. MiRNAs can be exported by cells through two main routes: (I) active transport via extracellular vesicles (EVs) and (II) transport as part of protein-miRNA complexes; which confer them protection from degradation by ribonucleases. Further, there can be some passive leakage of miRNAs from damaged cells (Figure 1) (13).
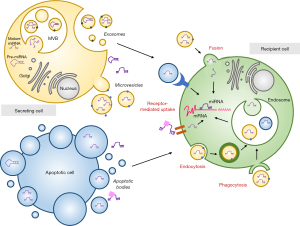
EVs are classified based on their size and biogenesis: exosomes (30–200 nm), microvesicles (MVs) (100–1,000 nm) and apoptotic bodies (ABs) (>1,000 nm). Their heterogeneity of cargoes includes lipids, proteins, metabolites and nucleic acids. Exosomes, also termed small EVs, are generated through the endocytic pathway through the translocation of multivesicular bodies (MVBs) to the plasma membrane, where they undergo fusion and release their contents through the process of exocytosis involving both endosomal sorting complex required for transport (ESCRT)-dependent or ESCRT-independent ceramide-mediated pathways (14). Their characteristic composition of surface proteins facilitates a selective targeting of recipient cells (15). The main proteins incorporated in exosomes are members of the tetraspanin family (CD9, CD63 and CD81), ESCRT proteins (Alix, TSG101), integrins, heat shock proteins (Hsp), actin and flotillins (14). A growing evidence indicates a selective active loading or sorting of miRNAs into these vesicles (16,17). Some studies suggest the involvement of AGO2 and other RNA-binding proteins in the regulation of this miRNA loading (18). Other RNA-binding proteins such as Y-box protein 1 (19), nucleophosmin 1 (NPM1) (20), neutral sphingomyelinase 2 (nSMase2) (21) and hnRNPA2B (22) also confer specificity to this process. The EXOmotif GGAG present in some miRNAs can be recognized by hnRNPA2B1, thus controlling the loading of these miRNAs. Interestingly, sumoylation of hnRNPA2B1 seems to be essential for the binding of hnRNPA2B1 to miRNAs (22). Recently, Garcia-Martin et al. identified sequence patterns in miRNAs which determine their secretion in EVs (EXOmotifs) or cellular retention (CELLmotifs), defining cell-type-specific EV miRNA profiles (23).
MVs and ABs are EVs formed by direct outward budding and fission of the plasma membrane in living and dying cells, respectively, and their surface protein largely depend on their cellular membrane of origin (24). MVs generation mostly occur in lipid-rich plasma membrane microdomains (lipid rafts/caveolae). Although the mechanism of miRNA uploading into MVs is largely unknown, Collino et al. demonstrated that the ribonucleoproteins T-cell internal antigen-1 (TIA), TIA-1-related (TIAR) and AU-rich element-binding protein (HuR) are involved in the selected miRNA pattern in MVs (25). Many MV-encapsulated miRNAs can also be associated with RISC proteins such as AGO2, which increase their stability and functionality in recipient cells (26). Phosphatidylserine is a distinctive element of ABs. Zernecke et al. firstly showed that miR-126-enriched ABs shed by endothelial cells could alter chemokine responses in neighboring cells (27). However, there are very few studies investigating the AB-encapsulated miRNA effects and a limited understanding about the specificity and selectivity of miRNA loading into ABs.
MiRNAs can also be transported in vesicle-free systems, such as RNA-binding proteins or low-density (LDL) and high-density lipoproteins (HDL) (28). More than half of the miRNAs found in serum may be bound to ribonucleoproteins, such as AGO2, NPM1 and ribosomal protein L10a and L5 (20,29). Of note, the profile of miRNAs bound to vesicle-free systems differs from that found in EVs, indicating complementary and independent mechanisms of miRNA transport (28).
Many research groups have demonstrated that extracellular miRNAs are functional in recipient cells. However, the mechanisms of miRNA uptake are not fully understood. There is evidence that vesicle-associated extracellular miRNAs can be internalized by recipient cells through endocytosis, phagocytosis or the direct fusion with the target-cell plasma membrane. Interaction can operate through two main mechanisms: receptor-ligand binding and direct release of EV content in target cells (Figure 1). Vesicle-free secreted miRNAs may be taken up by specific cell surface receptors. Particularly, miRNAs associated with HDL interact with the HDL receptor and scavenger receptor BI (SR-BI). MiRNAs have also been shown to be transferred via direct cell-cell contact and gap junctions (10).
The fact that many circulating miRNAs dynamically exhibit a bio-fluids-specific profile in relation to a pathophysiological state, not only constitutes a specific mechanism for intercellular communication, but also gives rise to the miRNA application as diagnostic and prognostic biomarkers (10). Particularly, miRNAs-based biomarkers in blood and urine have generated a strong interest in the field of nephrology. This review discusses the up-to-date knowledge of miRNA function in kidney diseases, focusing on their participation in cellular communication and their value as biomarkers. We present the following article in accordance with the Narrative Review reporting checklist (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-2/rc).
Methods
Bibliography search strategy includes publications in English from peer-reviewed journals listed in PubMed database from 2000–January 2022. Search terms used were: “extracellular miRNAs” OR “kidney disease” OR “biomarkers” (Table 1).
Table 1
| Criterion | Specification |
|---|---|
| Date of search | January 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | “Extracellular miRNAs” OR “kidney disease” OR “biomarkers” |
| Timeframe | 2000–January 2022 |
| Inclusion and exclusion criteria | Only articles written in English language and published in peer-reviewed journals were included |
| Selection process | Selection process was carried out by the author |
miRNAs, microRNAs.
Extracellular miRNAs in renal pathophysiology
Chronic kidney disease (CKD)
CKD is a clinical condition with maintained reduction of renal function which is present in 30–40% of patients with highly prevalent pathologies such as diabetes mellitus and hypertension (30). It leads to tubulointerstitial and glomerular fibrosis as a result of excessive deposition of extracellular matrix of proteins (ECM), such as hyaluronic acid, fibronectin (FN), proteoglycans and interstitial collagens in association with a persistent inflammatory response, tubular epithelial cell (TEC) dedifferentiation and loss and rarefaction of the peritubular microvasculature (30). Myofibroblasts, derived from fibroblasts and PDGFRβ+/PDGFRα+ mesenchymal cells in the kidney, are the principal cells responsible for producing ECM (31).
Regulation of kidney fibrosis by miRNAs
MiRNAs have raised as powerful dynamic regulators of fibrotic processes [fibromiRs, (32)]. It mainly occurs thought the regulation of transforming growth factor beta 1 (TGF-β1) signaling in a cell-dependent and context-dependent manner (33). In keeping, Dicer1 deficiency promotes fibrosis in different organs by upregulating Smad2/3 (34,35). TGF-β signaling, in turn, can regulate the transcription of miRNAs by binding Smad proteins to Smad-binding elements (SBEs) in the DNA. In addition, Smad-activated auxiliary factors such as the RNA helicase p68, a component of the Drosha microprocessor complex, can promote the recruitment of Drosha/DGCR8 to specific pri-miRNAs (36).
Gomez et al. identified 24 miRNAs commonly upregulated both in human CKD and in animal models of kidney fibrosis, suggesting a “fibrotic” miR signature in the kidney (37). Cell-specific small RNA-sequencing (sRNA-seq) on TECs, endothelial cells, PDGFR-β+ cells and macrophages also show a differential miRNA profile in injured kidneys (38).
MiRNA-mediated control of kidney fibrosis mainly occurs thought the regulation of crucial signaling pathways associated with epithelial dedifferentiation, myofibroblast activation, matrix deposition and inflammation (Table 2) (113). MiR-21 regulates TGF-β-induced signaling pathways by targeting Smad7 and phosphatase and tensin homology (PTEN) and, in turn, is upregulated by TGF-β in TECs promoting renal fibrosis (39,40). Other miRNAs regulating TGF-β signaling are: miR-433 which contributes to renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway (42), miR-23b targeting TGF-β receptor type II (TGF-βRII), SMAD3 and TGF-β, suggesting a negative feedback loop-regulating TGF-β signaling (43), and miRNA-196a/b, which mitigate renal fibrosis by targeting TGF-βRII (44). Although the contribution of epithelial to mesenchymal transition (EMT) to fibrosis is seriously questioned (31), some miRNAs have been closely related to it. MiR-200a targets include EMT-related factors such as TGF-β2, β-catenin and the Zinc finger E-box binding homeobox 1 (ZEB1), which negatively modulates E-cadherin (45-47). Let-7d and miR-214 has been described as regulators of key EMT genes SNAIL and TWIST (48-50). Metabolic derangement, particularly mitochondrial impairment in TECs, is now identified as a key culprit in fibrogenesis (114). MiR-21 promote fibrosis by silencing metabolic pathways targeting peroxisome proliferator-activated receptor alpha (PPAR-α) and MPV17-like protein (39). MiRNA-27a also promotes fibrosis via suppressing PPAR-γ pathway (52), while disruption of mitochondrial oxidative phosphorylation during CKD is also a consequence of miR-214 increase, by targeting the mitochondrial genes MT-ND6 and MT-ND4L (51). Impairment of renal fatty acid oxidation during fibrosis progression is a process controlled by miR-33 (115), miR-150 and miR-495 (53). Other miRNAs can regulate ECM production during CKD. TGF-β/Smad3 signaling inhibits miR-29 in TECs which targets collagens, fibrillins, laminins and elastin (55). MiR-21 also has a role in ECM homeostasis through the regulation of metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) (41). MiRNA-132 and miR-503 can reduce renal fibrosis by selectively inhibiting myofibroblast proliferation (56,57).
Table 2
| miRNAs | Level during disease | Target | Effect | Reference(s) |
|---|---|---|---|---|
| Chronic kidney injury | ||||
| miR-21 | Up | SMAD7, PTEN, PPARA, MMPs, TIMPs | ECM*, mitochondrial dysfunction* | (39-41) |
| miR-433 | Up | AZIN1 | ECM* | (42) |
| miR-23 | Up | TGFBRII, SMAD3, TGFB | ECM# | (43) |
| miR-196 | Up | TGFBRII | ECM# | (44) |
| miR-200a | Up | TGFB2, CTNNB1, ZEB1/2 | EMT# | (45-47) |
| Let-7 | Down | TGFBRI, HMGA2 | EMT* | (48,49) |
| miR-214 | Up | SNAIL, TWIST, ND6, ND4 | EMT* | (50,51) |
| miR-27 | Up | PPARG | Mitochondrial dysfunction* | (52) |
| miR-33/-150/-495 | Up | CPT1A | Mitochondrial dysfunction* | (53,54) |
| miR-29 | Up | COLs, FBNs, LMNAs, ELNs | ECM# | (55) |
| miR-132 | Up | FOXO3A, P300 | Fibroblast proliferation# | (56) |
| miR-503 | Down | RAF1 | Fibroblast proliferation# | (57) |
| Acute kidney injury | ||||
| miR-21 | Up | PDCD4, PTEC, PPARA, NFKB | Apoptosis#, inflammation# | (58,59) |
| miR-494 | Up | ATF-3 | Apoptosis*, inflammation* | (60) |
| miR-24 | Up | H2A.X, HO-1 | Apoptosis* | (61) |
| miR-194 | Down | RHEB | ROS#, inflammation# | (62) |
| miR-181 | Up | BCL-2 | Apoptosis* | (63) |
| miR-489 | Up | PARP1 | Apoptosis# | (64) |
| miR-150 | Up | MYB | Apoptosis*, inflammation* | (65) |
| miR-16 | Up | BCL2 | Apoptosis* | (66) |
| miR-107 | Up | DUSP7 | Inflammation* | (67) |
| miR-183 | Up | SIRT1 | Fibrosis*, apoptosis* | (68) |
| Diabetic nephropathy | ||||
| miR-192 | Up | ZEB1/2 | ECM* | (69) |
| miR-200b/c | Up | ZEB1, FOG2 | ECM* | (70) |
| miR-216/-217 | Up | PTEN | Cellular hypertrophy* | (71) |
| miR-21 | Up | SMAD7, MMP-7, TIMP1 | ECM* | (41,72) |
| miR-23 | Down | SNON | ECM*, EMT* | (73) |
| miR-29a | Down | HDAC | Podocyte damage* | (74) |
| miR-29c | Up | SPRY-1 | Apoptosis*, ECM* | (75) |
| miR-25 | Down | NOX4 | ROS* | (76) |
| miR-30/-130b | Up | SNAIL1, CTGF, GIPR2 | EMT# | (77-79) |
| miR-10 | Up | NLRP3 | Inflammation# | (80) |
| miR-45 | Up | PSHD11, LMP7, P65 | Inflammation# | (81,82) |
| miR-377 | Up | P21, Mn-SOD | ROS*, autophagy# | (83) |
| miR-214 | Up | ULK1, PTEN | Autophagy#, ECM*, cellular hypertrophy* | (84,85) |
| miR-150 | Up | SIRT1 | Autophagy# | (86) |
| Hypertensive nephropathy | ||||
| miR-29 | Up | COL1A1 | ECM# | (87) |
| miR-204 | Down | SHP2 | ECM* | (88) |
| miR-192 | Up | ZEB1/2 | ECM* | (89) |
| miR-155 | Up | AGTR1 | ECM# | (90) |
| miR-21 | Up | PPARA | ECM* | (91) |
| miR-103 | Up | SNRK | ECM*, inflammation* | (92) |
| miR-429 | Up | ZEB1 | EMT# | (93) |
| Kidney immune diseases | ||||
| Let7a miR-148/-196 | Up | GALNT2, C1GALT1 | Aberrant IgA glycosylation* | (94-96) |
| miR-223 | Down | KPNA3/1 | Endothelial cell proliferation* | (97) |
| miR-100 | Down | IL-8 | Inflammation* | (98) |
| miR-877 | Down | IL-1β | Inflammation* | (98) |
| miR-200bc miR-429 |
Up | TWEAK | Inflammation# | (99) |
| miR-21 | Up | PTEN | ECM* | (100) |
| miR-146 | Up | TRAF6 | Inflammation# | (101) |
| miR-150 | Up | SOCS1 | ECM* | (102) |
| miR-422 | Up | KLK4 | ROS*, inflammation* | (103) |
| miR-10 | Down | IL-8 | Inflammation* | (104) |
| Polycystic kidney disease | ||||
| miR-17 | Up | PKD1/2, PPARA | Cyst growth* | (105,106) |
| miR-92 | Up | HNF1B | Cyst growth* | (105) |
| miR-20/-106a | Down | KLF12 | Cell proliferation* | (107) |
| miR-365 | Up | PKHD1 | ECM# | (108) |
| miR-192/-194 | Down | ZEB2, CADH2 | EMT* | (109) |
| miR-21 | Up | PDCD4 | Apoptosis# | (110) |
| miR-199a | Up | CDKN1C/P53 | Cell proliferation* | (111) |
| miR-214 | Up | TLR4 | Inflammation# | (112) |
* and # represent enhanced or repressed process, respectively. miRNA, microRNA; ECM, extracellular matrix; EMT, epithelial-mesenchymal transition; ROS, reactive oxygen species.
Extracellular miRNAs in renal fibrosis: potential biomarkers
In the kidney, EVs can be originated from all segments of the nephron and have an important role as signaling messengers, regulating the phenotype of renal and extrarenal cell types controlling various biological processes including programmed cell death, angiogenesis, inflammation, immunosuppression and regeneration, and subsequently, the outcome of kidney injury (116).
Crosstalk between kidney cells, including injured and uninjured TECs, fibroblasts and immune cells, has emerging as a crucial mechanism during fibrogenesis which involves EVs shuttled miRNAs (Figure 2) (117). TGF-β1 mRNA is secreted by injured TECs and transported to interstitial fibroblasts through exosomes (118). Hypoxia-injured proximal TECs (PTECs) promote fibroblast activation by shuttling exosomes containing miR-150 (119). In fibrotic tissue, secreted miR-21-containing MVs from injured and senescent TECs promote EMT in neighboring cells by targeting PI3K-Akt and PPARα-HIF-1α pathways, respectively (120-122), while MV-secreted miR-216a promotes EMT and aggravates renal fibrosis through the PTEN/Akt pathway (123). By contrast, miR-34a is secreted by interstitial fibroblasts and transported via MVs toward TECs, inducing apoptosis and tubular atrophy (124).

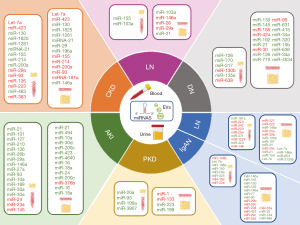
Urine and serum levels of some fibromiRs correlate with proteinuria and kidney function in CKD patients (Figure 3). Muralidharan et al. provided an extracellular miRNA signature of 384 urinary and 266 circulatory miRNAs differentially expressed between patients with estimated glomerular filtration rate (eGFR) ≥30 versus <30 mL/min/1.73 m2. Thus, let-7a and miR-423 showed lower levels in urine and plasma of patients with eGFR <30, respectively while the presence of miR-130, miR-1825 and miR-1281 was upregulated in urine and plasma of patients with eGFR <30; which was confirmed in albumin/TGF-β1-treated mice and TECs (125). Increased serum and urinary miRNA-21 levels, the best characterized miRNA in CKD, parallel the severity of kidney fibrosis and renal loss of function (126,127). In another cohort of CKD patients, increased miR-29 and miR-196a and decreased miR-155, miR-214, miR-200a and miR-93 levels were observed in urine, while increased miR-155, miR-214 and miR-200a and reduced miR-29a and miR-93 were found in serum of CKD patients, suggesting their use as potential non-invasive disease biomarkers (128). Another study examined miR-126 and miR-223 in serum of 601 CKD patients with a follow-up for 6 years, noticing that the level of both miRNAs had decreased along time (129). Serum miR-483 and miR-363 were identified by next generation sequencing as potential diagnostic biomarkers associated with CKD severity (130). By the same approach, Khurana et al. identified 30 differentially expressed ncRNAs in urinary exosomes from CKD patients. Among them, miRNA-181a was 200-fold decreased and appeared as the most robust and stable potential biomarker (131). Ichii et al. found increased levels of miR-146a in urine sediments of mice with CKD (132).
Acute kidney injury (AKI)
AKI is characterized by the rapid decline of renal function due to ischemia, nephrotoxicity, sepsis, obstruction of urinary tract or bladder outflow obstruction which can lead to tubular necroinflammation. AKI is a predisposing cause and an occasional precedent of CKD, whereas CKD is a major risk factor for AKI (133).
Regulation of AKI by miRNAs
The importance of miRNAs in AKI was first evidenced by the conspicuous protective effect observed in PTEC-specific Dicer knockout mice where over 80% miRNAs were depleted (134). Altered expression of >50 miRNAs have been described to play protective and pathogenic roles in the development of AKI including miR-21, miR-205, miR-127 and miR-494 (60,135-137). Cell-enriched miRNA analysis in AKI shows macrophage-enhanced miR-18a and miR-16 and PTEC-enhanced miR-194 (38).
Inflammation and apoptosis are the main responses regulated by miRNAs during AKI (Table 2). MiRNA-21 inhibits apoptosis by targeting programmed cell death protein-4 (PDCD4) and PTEN (138), while showing an anti-inflammatory role by targeting PPAR-α and nuclear factor-kappa B (NF-κB) (58,59). MiR-494 is up-regulated in AKI repressing activating transcription factor-3 (ATF-3) which increases inflammatory mediators (60). In turn, miR-219 downregulation triggers a proinflammatory phenotype of macrophages increasing the expression of Mincle during AKI (139). Other miRNAs such as miR-24 (61), miR-181 (63), miR-150 (65), miR-16 (66) and miR-183 (68) stimulate apoptosis during AKI, while miR-489 is induced via hypoxia-inducible factor-1 (HIF-1α) to protect from apoptosis (64). Endothelial activation also plays a key role in septic AKI, involving altered vascular reactivity, permeability and adherence of leukocytes. It has been described that increased miR-107 induces tumor necrosis factor alpha (TNF-α) secretion by targeting dual-specificity phosphatase 7 (DUSP7) in endothelial cells (67).
Extracellular miRNAs in AKI: potential biomarkers
EV-borne miRNAs also participate in cell-to-cell communication during AKI (Figure 2). Hypoxia-injured PTECs trigger the pro-inflammatory phenotype of macrophages by exosomal miR-23a and miR-19b transferring which suppress the activity of the ubiquitin editor A20 and NF-κB/SOCS1, respectively (140,141). MiR-20a is enriched in hypoxia-derived tubular exosomes and protects against TECs mitochondrial failure and apoptosis (142). In turn, miR-191 secreted by platelet-derived MVs induces apoptosis of TECs in AKI by targeting cystathionine-β-synthase (CBS) (143).
There is an urgent need to identify sensitive and specific early markers for AKI that overcomes the limitations of traditional serum creatinine and blood urea nitrogen values which change only after significant kidney injury with a substantial time delay. Some serum and urine miRNAs have raised as promising early indicators of this disease (Figure 3) (144). It has been described that increased serum and urinary miRNA-21 levels parallel the severity of AKI (145). Other miRNAs with differential urine levels are miR-494, miR-10a, miR-30d, miR-200c, miR-423 and miR-4640 (60,146,147). MiRNA profiling of urinary exosomes shows that miRNA concentration tracks with AKI progression, including miR-16, miR-24 and miR-200c. Complementarity, miR-9a, miR-141, miR-200c and miR-429 were associated to AKI recovery stage (148). Several miRNAs, including miR-101, miR-127, miR-210, miR-126, miR-26b, miR-29a, miR-146a, miR-27a, miR-93 and miR-10a have also been reported to be altered in serum of AKI patients (149). Noteworthy, miR-210 predicted AKI mortality in intensive care unit patients (150).
Specific serum and urine miRNA signatures have been associated with the different casualties of AKI. Pavkovic et al. detected more than 20-fold changes in 11 urinary miRNAs of the cisplatin-induced AKI model, which are associated with DNA damage response, cell cycle dysregulation and apoptosis (151). In different models of contrast-induced AKI, increased miR-188, miR-30a and miR-30e levels are detected in plasma (152). Decreased urinary miR-376b is proposed as a useful biomarker for the diagnosis or identification of AKI in patients with sepsis. Mechanistically, miR-376b is suppressed by NF-κB in TECs, leading to the induction of its target gene NF-κB inhibitor zeta (NFKBIZ), which limits inflammation and cell death (153). A significant upregulation of miR-16 and miR-18a was observed in the first-passed urine of patients who developed delayed graft function (DGF), the typical first clinical AKI manifestation occurring following renal transplantation (38). Decreased serum miR-24, miR-23a and miR-145 levels were reported in post-myocardial infarction AKI pathogenesis (154).
Diabetic nephropathy (DN)
DN is a complication of type 1 and 2 diabetes mellitus, with a global incidence of 9%. It is histologically characterized by early tubular cell atrophy followed by mesangial cell (MCs) hypertrophy, podocyte dysfunction, glomerulosclerosis, renal fibrosis and matrix expansion. DN main contributors are hyperglycaemia and insulin resistance (155).
Regulation of DN by miRNAs
First evidences of the involvement of miRNAs in DN were observed in mice with podocyte-specific deletion of Dicer or Drosha, which exhibited severe renal phenotypes including proteinuria, podocyte foot process effacement and apoptosis, glomerulosclerosis and tubulointerstitial fibrosis (Table 2) (156,157).
MiR-192 is a master miRNA regulator of DN (158). MiR-192 upregulates collagen genes in MCs by targeting the transcriptional repressors ZEB1/2 (69). Of interest, miR-192 upregulates other miRNAs, including miR-216a/miR-217 and miR-200b/c whose targets promote cellular hypertrophy (71). MiR-21 and miR-23 also promote renal fibrogenesis and hypertrophy by regulating TGF-β signaling (41,73). Inflammation is an early event during DN. MiR-10 negatively regulates inflammation by targeting activation of the NLRP3 inflammasome (80), while miR-45 downregulates the 26S proteasome non-ATPase regulatory subunit 11 (PSMD11), multifunctional protease 7 (LMP7) and NF-κB p65 (81,82). Redox and autophagy imbalance are also drivers of DN. Enhanced expression of miR-377 increases FN levels by targeting p21-activated kinase and manganese superoxide dismutase (MnSOD) (83). MiR-214 and miR-150 control autophagy during DN by regulating the PI3K/AKT/mTOR and SIRT1/p53/AMPK pathways, respectively (84,85). Other miRNAs modulate the high-glucose (HG)-induced homeostasis imbalance in DN. HG induce apoptosis of mouse podocytes by downregulating miR-29a and increasing miR-29c expression. Low levels of miR-29a correlate with enhanced levels of its target histone deacetylase (HDAC), which modulates the acetylation status of nephrin (74). MiR-29c promotes apoptosis and FN synthesis by inhibition of sprouty RTK signaling antagonist 1 (SPRY1) (75). Other miRNAs involved in the HG-induced response are miR-25 (76), miR-30c and miR-130b (77-79), which modulate reactive oxigen species (ROS) and EMT, respectively.
Extracellular miRNAs in DN: potential biomarkers
EV miRNAs also participate in the cellular crosstalk during DN (Figure 2). EVs enriched in miR-196b from TECs mediate fibroblast activation and promotes renal fibrosis in diabetic mice through the activation of the signal transducer and activator of transcription 3 (STAT3)/suppressor of cytokine signaling 2 (SOCS2) signaling pathway (159). MiR-199a is increased in EVs from human serum albumin (HSA)-induced TECs which triggers kidney macrophage M1 polarization by targeting the Klotho/toll like receptor 4 (TLR4) pathway (160). EVs enriched in miR-4756 are also produced by HSA-induced TECs and promote EMT and endoplasmic reticulum stress by targeting Sestrin2 (161). EVs from HG-stimulated TECs favors renal fibrosis by transferring miR-192 to healthy recipient TECs and targeting glucagon-like peptide 1 receptor (GLP1R) (162). EVs from injured podocytes containing miRNA-424 and miR-149 induce apoptosis and p38 phosphorylation in TECs (163).
Many of the altered urinary miRNAs in type 1 DN patients are associated renal fibrosis-associated pathways (Figure 3) (164). Urinary exosomes from diabetic patients with microalbuminuria are enriched in miR-130 and miR-145 while have reduced miR-155 and miR-424 levels (165). Urine miR-192 and miR-21 are significantly increased while miR-30b levels are decreased in diabetic patients with altered kidney function (166-169). Interestingly, miR-415 was found to be elevated in urinary exosomes prior to albuminuria and glomerulosclerosis which suggest its use as an early biomarker (170). Park et al. identified miR-126 and the miR-770 family in urine and blood as promising predictive markers of DN progression (171). Serum miR-217 levels positively correlates with severity of diabetes nephropathy (172). By contrast, reduced serum miR-130b correlate with it (173). Urinary sediment miR-95 and miR-631 also reflect the severity and prognosis of DN (174). MiR-135a levels are upregulated in serum and renal tissue from patients with DN and db/db mice, and correlate with microalbuminuria and renal fibrosis scores (175). MiR-34a and miR-320c are upregulated in the urinary exosomes of type 2 DN (176,177). MiR-34a regulates MC proliferation and glomerular hypertrophy by targeting growth arrest-specific 1 (GAS1) (178), while miR-320c protects TECs from damage by downregulating Bone Morphogenetic Protein 6 (BMP6) (179). Other potential biomarkers for type 2 DN are miR-15b, miR-636, miR-34a and miR-4534 in urine (177,180) and miR-638 in serum (181).
Hypertensive nephropathy
Hypertension affects 25–30% population worldwide. Under chronicity, elevated blood pressure damages renal vessels and impairs GFR, promoting nephropathy characterized by renal fibrosis, tubular and glomerular hypertrophy. In turn, about 80% of CKD patients eventually develop hypertension as a consequence of an unbalanced renin-angiotensin-aldosterone system (RAS) (182).
Regulation of hypertensive nephropathy by miRNAs
The miRNA profile of human hypertensive nephrosclerosis biopsies revealed differential expression, showing higher levels of miR-200a, miR-200b, miR-141, miR-429, miR-146, miR-132, miR-192 and miR-205 (183,184).
High salt intake, one of the risk factors of hypertension, increases miR-29b expression in renal medulla in non-salt sensitive rats. Due to several collagen genes are miR-29b target genes, its increase may have a protective role (Table 2) (87). In the same model, miR-204 is downregulated and promotes the protein tyrosine phosphatase nonreceptor type 11 (SHP2)/p-STAT3 signaling, exacerbating renal interstitial fibrosis (88). Angiotensin (Ang) II, the main peptide of the RAS, can exert both a vasoconstrictor effect and a pro-inflammatory action in post-glomerular arteries (185). MiR-155 controls blood pressure by downregulating the expression of the Ang II type 1 receptor (AGTR1) (90). In the same vein, miR-21 mediates Ang II-induced kidney fibrosis via amplifying the TGF-β1/Smad3 pathway by targeting PPAR-α (91), while miR-103a contributes to Ang II-induced renal inflammation and fibrosis through the SNF related kinase (SNRK)/NF-κB/p65 regulatory axis (92).
Extracellular miRNAs in hypertensive nephropathy: potential biomarkers
Gildea et al. identified 45 urinary exosomal miRNAs associated with salt sensitivity or inverse salt sensitivity (Figure 3) (186). Upregulated expression of miR-155 has been reported in plasma of renal transplant recipients (187). In patients with hypertensive nephropathy, miR-103a was upregulated in urine and serum. Interestingly, patients with a positive response to angiotensin-converting enzyme inhibitor or β-blocker treatment displaying a reduction in their albumin–creatinine ratio showed a decrease in miR-103a, suggesting this miRNA as a dynamic biomarker reflecting the pathological status and treatment response (188). MiR-146a, miR-26 and miR-29a from urinary exosomes correlates with albuminuria in essential hypertension (189-191), while higher urinary miR-21 levels are detected earlier than urinary albumin (184).
Kidney immune diseases
IgA nephropathy (IgAN) and lupus nephritis (LN) are the main kidney immune diseases. IgAN is the most common form of primary glomerulonephritis. It is characterized by the aberrant glycosylation of immunoglobulin A (IgA1) and consequently alters the synthesis and binding of antibodies directed against this IgA1 form, generating an immune complex that accumulates in the glomerular mesangium. It activates MCs, enhancing proliferation and secretion of ECM, cytokines and chemokines and leading to renal injury (192). LN is an autoimmune disorder with a complex pathophysiology whose immunological hallmark is the production of a range of autoantibodies directed at ubiquitous nuclear components (193).
Regulation of IgAN by miRNAs
MiRNA expression profiling of kidney biopsies from patients with IgAN showed that dysregulated levels of miRNAs related to fibrosis (downregulation of miR-200c and upregulation of miR-192, miR-141 and miR-205) (194) and to the immune response (upregulation of miR-155 and miR-146a) (195), associated with disease severity and progression. MiR-150 has been proposed as a potential functional mediator of kidney fibrosis progression in IgAN (196). Let-7a and miR-148b/miR-196b control N-acetylgalactosaminyltransferase 2 (GALNT2) and 1β,1,3 galactosyltransferase 1 (C1GALT1), respectively, enzymes which are involved in aberrant IgA glycosylation (94-96).
MiRNAs regulates IgA features in different renal cell types (Table 2). Glomerular endothelial cells of patients with IgAN showed decreased levels of miR-223, contributing to cellular proliferation. This miRNA targets importin α4 and α5 (KPNA3/1), responsible for the nuclear transport of NF-κB p65 and STAT3 (97). Downregulation of miR-100 and miR-877 controls overproduction of interleukin 8 (IL-8) and IL-1β in MCs activated by secretory IgA from IgAN patients (98). By contrast, miR-200bc/429 cluster alleviates inflammation in podocytes from IgAN patients by targeting TNF-related weak inducer of apoptosis (TWEAK)/FN14 (99). Of note, miR-21 promotes fibrogenic activation in podocytes and TECs by activating the PTEN/Akt pathway activation in IgAN (100).
Extracellular miRNAs in IgAN: potential biomarkers
Several studies have described the serum and urine miRNome of IgAN patients postulating some miRNAs as potential biomarkers (Figure 3). Serum miR-148b and miR-let-7b levels were reported to discriminate patients with IgAN from both controls and patients with other forms of glomerulonephritis (197). Plasma content in miR-148a, miR-150, miR-20a and miR-425 is increased in IgAN patients, especially in the early-stage (198), while IgAN patients with lower miR-192 levels are more likely to have renal function decline (199). Elevated urinary levels of miRNA-146a and miRNA-155 in IgAN patients, correlate with proteinuria but inversely correlate with urinary expression of the cytokines IL-1β and TNF-α (195). Other potential biomarkers of IgAN are miR-150, miR-155, miR-146a, miR-17 and miR-93—which are found at increased levels—and miR-29b, miR-29c, miR-204, miR-431 and miR-555—which are found at decreased levels—in the urine of patients with IgAN (200,201). These changes have also been reported in miR-29c and miR-146a urinary exosomes (202). Levels of miR-33a both in serum, urine and kidney tissues decrease with the severity of renal injury and the progression of renal failure in IgAN patients (54). Increased levels of miR-25, miR-144 and miR-486 in urinary sediment, mainly derived from urinary erythrocytes, could also be non-invasive candidate biomarkers for IgAN (203).
Regulation of lupus nephritis by miRNAs
MiRNA expression evaluation in kidney biopsies from class II LN patients (characterized by pure mesangial involvement), identified 36 upregulated and 30 downregulated miRNAs (204). In patients with LN, disease severity correlates and glomerular and tubulointerstitial expression of miR-638, miR-198 and miR-146a (205). MiR-150 has also been demonstrated to promote renal fibrogenesis by targeting the suppressor of cytokine signaling 1 (SOCS1), a negative regulator of the JAK/STAT signaling pathway, which controls cell proliferation, inflammation and fibrosis (Table 2) (206). MCs contribute to the pathogenesis of LN through both the secretion of proinflammatory cytokines and matrix protein deposition. MiR-422a is upregulated in MCs and TECs from renal LN biopsies. This miRNA targets kallikrein related peptidase 4 (KLK4) which belongs to the kallikrein-kinin system and have pleiotropic effects in inflammation, oxidative stress and vascular function (103). IL-8, essential in type III hypersensitivity and a major characteristic of LN, was confirmed as a direct target of miR-10a in MCs (104).
Extracellular miRNAs in lupus nephritis: potential biomarkers
Some miRNAs have emerged as potential LN biomarkers (Figure 3). MiR-181a was increased while miR-223 was decreased in serum samples from LN patients, whose levels correlate with markers of this disease (68). MiRNA expression profiles in plasma, urinary sediment and peripheral blood mononuclear cells (PBMCs) revealed a group of miRNAs associated with LN (miR-342, miR-223 and miR-20a in plasma, miR-221 and miR-222 in urinary sediment, and miR-371, miR-1224 and miR-423 in PBMCs), which are promising disease biomarkers (207-209). Although, miR-146 expression is increased in the glomerulus, the levels are decreased in PBMCs from patients with LN. However, miR-146 enrichment in urinary exosomes associates with renal damage and can discriminate patients with active from inactive LN patients (210). Levels of miR-26a and miR-30b are decreased in the kidneys and urine of LN patients and promote MC proliferation (211). Urinary exosomal miR-135b, miR-107 and miR-31 levels are higher in treatment responder patients. These are mainly produced in TECs and are engulfed by endothelial and MCs where target HIF-1α, reducing proliferation and inflammation (212). Decreased urinary exosome-associated miR-29c appears to be a predictor of early renal fibrosis in LN (213), while in the case of miR-21, miR-150 and let-7a correlate with the clinical stage (214). MiR-146a, miR-654 and miR-3135b in urinary exosomes possess a predictive value in type IV LN complicated by cellular crescent (215).
Polycystic kidney disease (PKD)
PKD is a genetic kidney disorder characterized by the growth of cysts in the kidneys, due to mutations or dysregulated expression of either polycystic kidney disease 1 (PKD1) or 2 (PKD2) gene, which encode for polycystin 1 and 2, respectively, with an autosomal dominant (ADPKD) pattern of inheritance, or the polycystic kidney and hepatic disease 1 (PKHD1) gene which encodes for fibrocystin/polyductin, resulting in an autosomal recessive (ARPKD) disease. Aberrant expression of these genes leads to disrupted cell division, proliferation and impaired cell-matrix and/or cell-cell interactions (216).
Regulation of PKD by miRNAs
Genetic profiling assays revealed alterations in miRNA expression profiles in PKD, most of them directly regulating the expression of PKD1, PKD2, PKHD and cell proliferation-related genes (Table 2) (217). Thus, the dysregulated miRNAs miR-214, miR-31, miR-199a, miR-21, miR-34a, miR-132 and miR-146b are believed to target major pathways in autosomal dominant PKD (218). Pandey et al. predicted and verified several miRNAs (miRs-10a, miR-30a, miR-96, miR-126, miR-182, miR-200a, miR-204, miR-429 and miR-488): mRNA reciprocal interactions in a PKD mouse model (217).
The miR-17~92 cluster is upregulated in the kidney of various PKD mouse models and its overexpression produces cysts. Particularly, miR-17 targets PKD1 and PKD2 whereas miR-92 inhibits PKHD1 through the transcription factor hepatocyte nuclear factor 1 homeobox B (HNF-1β). It has been described that the cellular myelocytomatosis oncogene (C-MYC) transcriptionally activates the miR-17~92 cluster, which regulates the mechanistic target of rapamycin kinase (mTOR) and TGF-β pathways, both of which are implicated in cyst growth (105). Tran et al. demonstrated that the binding of miR-17 to PKD2 is antagonized by the RNA-binding protein bicaudal C homolog 1 (BICC1), thus regulating PKD2 gene dosage (219).
MiRNAs have a key role in cyst expansion which is associated with EMT. PKHD1 is a direct target of miR-365 and is involved in the decrease of E-cadherin and destruction of ECM (108). MiR-192 and miR-194, whose levels are downregulated by hypermethylation of their promoter region, directly target ZEB2 and cadherin-2, which are involved in EMT. MiR-199a inhibitor suppresses proliferation of cystic cells and induces cell apoptosis by targeting cyclin dependent kinase inhibitor 1C (CDKN1C)/p57, a negative regulator of cell proliferation by inhibiting G1 cyclin-dependent kinases (111).
Extracellular miRNAs in PKD: potential biomarkers
The role of EVs in ADPKD progression have not drawn considerable attention so far (Figure 3). Of note, EVs generated from cystic TECs display increased levels of miR-200b, miR-200c, miR-429 and miR-21 levels which promote cyst growth in ADPKD by inducing epithelial cell proliferation, fibroblast activation and macrophage recruitment (220).
The serum and urine miRNA profile in ADPKD patients differ substantially depending on the stage of CKD. Serum exosomal levels of miR-17 family members miR-20a, miR-93 and miR-106a significantly drop after hemodialysis in ADPKD patients (221). In urine from ADPKD patients, dysregulated miRNAs have previously involved in kidney tumor suppression (miR-1 and miR-133). Others miRNAs have presumed inflammatory and fibroblast cell origin (miR-223/miR-199) (222). Increased levels of miR-3907 in the circulation can predict ADPKD progression (223).
Clinical applications and perspectives
Since early 1940s, the mainstay of renal functional monitoring has not progress significantly. So, the increased estimated prevalence of kidney diseases urgently demands novel biomarkers to enhance early diagnosis, guide prognosis and monitor response to treatment (224). The importance of miRNAs in the kidney field has gained widespread interest over the last decade not only enabling to understand in-depth the pathways involved in kidney pathophysiology, but also as biomarkers circulating in biofluids. In kidney diseases, urine is the preferred source of biomarkers due to its direct access to the damaged tissues of the kidney and urinary tract. Although a kidney disease-specific blood and urine miRNA signature has been identified in patients and mouse models, no definitive data have yet been translated to the clinic (225). There is limited knowledge about the cell type of origin and functional role of circulating miRNA and therefore, if they are tissue/disease-specific or represent more general pathologies like inflammation. Understanding the cell specificity of miRNAs’ expression in the kidney is essential to contextualize whole-tissue miRNA changes and target validation, which would improve the rational selection of biomarkers. Identification of host cell-derived protein surface markers among EVs surface proteins would allow to determine their cellular origin (226). Further, unification of methodology for extracellular RNA isolation, purification and detection by combining-omics technologies, including proteomic, transcriptomic, and metabolomic methods, as well as to standardize EVs classification is needed to provide more robust biomarkers. Consideration of the distinct miRNA half-lives, conservation degree between species and variability between patients, evaluating variables such as age, gender, ethnicity, additional medication or the presence of comorbidities in large cohort studies are also essential steps for their clinical translation (227).
While a miRNA-based therapy that either restores or blocks miRNA expression and activity by using miRNA mimics or antagomirs is very appealing, the potential of miRNAs as an effective therapy has been limited so far to experimental models and two ongoing clinical trials (clinicaltrials.gov, NCT03373786, NCT04536688), mainly due to the lack of reliable organ- and cell-specific delivery methods and off-target effects on other genes. Some new synthesis and delivery methodology include locked nucleic-acids (LNA) (228) coupled to lipid-based nanoparticles, FDA-approved poly-lactic-co-glycolic acid (PLGA)-based nanoparticles, cell and/or tissue-specific antibodies/peptides and ultrasound microbubble-mediated gene transference (229,230). EVs have also gained prominence as drug delivery vehicles. The process of loading EVs with specific cargos, including miRNA analogues, can be attained by manipulating parental cells (endogenous loading) or the isolated EVs (exogenous loading) themselves. Furthermore, to boost their delivery and biodistribution, EVs can be engineered to recognize specific cell surface receptors (231). How to obtain exosomes on a large scale for clinical treatment will also be a focus of future studies. In this line, several studies describe the renoprotective effect of EVs derived from multipotent mesenchymal stem cells (MSCs), emerging as a potential powerful cell-free therapeutic strategy. In some kidney diseases, content analysis of this MSC-derived EV identified that transported miRNAs can underlie this response (Table 3) (246). Future studies are directed to integrate circulating and renal miRNA expression data along renal cell types to generate a complete kidney expression atlas in injury and repair conditions coupled to ligand-receptor networks between EVs and kidney cells, as well as to develop new technology for single-vesicle analysis, which undoubtedly will unleash their full potential as therapeutic targets and biomarkers.
Table 3
| MSC source | In vivo model | miRNA | EV subtype | Pathophysiological effects | Target gene | Reference(s) |
|---|---|---|---|---|---|---|
| Bone marrow | UUO | miR-144 | Exosomes | Tubular basement membrane integrity | tPA | (232) |
| Bone marrow | UUO | Let-7c | Exosomes | ECM accumulation | TGFBRI | (233) |
| Muscle | UUO | miR-29 | Exosomes | ECM accumulation | TGFB3 | (234) |
| Endothelium | IRI | miR-126, miR-296 | Microvesicles | Capillary rarefaction, glomerulosclerosis, tubulointerstitial fibrosis | SPRED1, VCAM1, PIK3R2, HGS | (235) |
| Epithelium | Glycerol | miR-10a, miR-486, miR-127 | Exosomes | Necrosis | Non-described | (236) |
| Umbilical cord | CLP | miR-146b | Exosomes | Inflammation, apoptosis | IRAK1 | (237) |
| Bone marrow | IRI | miR-199a | Exosomes | Apoptosis | SEMA3A | (238) |
| Non-described | Cisplatin | miR-1184 | Exosomes | Inflammation, apoptosis | FOXO4 | (239) |
| hWJMSCs | IRI | miR-30b/c/d | EVs | Mitochondrial fission, apoptosis | DRP1 | (240) |
| ECFC | IRI | miR-486 | Exosomes | Apoptosis | PTEN | (241) |
| Placenta | IRI | miR-200a | EVs | Oxidative stress, mitochondrial fragmentation | KEAP1 | (242) |
| Umbilical cord | STZ | miR-451 | MVs | Apoptosis, EMT, ECM accumulation | P15, P19 | (243) |
| Bone marrow | STZ | Let-7a | Exosomes | Apoptosis, oxidative stress, EMT, ECM accumulation | USP22 | (244) |
| Adipose | db/db | miR-26 | Exosomes | Apoptosis | TLR4 | (245) |
The treatment with MSC-EVs reduces the pathophysiological effects indicated in mouse models of chronic (UUO) and acute (IRI, glycerol, CLP, cisplatin) kidney disease and diabetic nephropathy (STZ, db/db transgenic mouse), respectively. MSC, mesenchymal stem cell; EV, extracellular vesicle; miRNA, microRNA; UUO, unilateral ureteral obstruction; ECM, extracellular matrix; IRI, ischemia reperfusion injury; CLP, cecal ligation and puncture; STZ, streptozotocin. MV, microvesicle; EMT, epithelial-mesenchymal transition.
Acknowledgments
Funding: VM is supported by a FEBS Long-Term Fellowship.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Chenyu Zhang) for the series “Extracellular RNAs and Human Health” published in ExRNA. The article has undergone external peer review.
Reporting Checklist: The author has completed the Narrative Review reporting checklist. Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-2/rc
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-2/coif). The series “Extracellular RNAs and Human Health” was commissioned by the editorial office without any funding or sponsorship. VM reports a long-term fellowship from Federation of European Biochemical Societies, which covers her postdoc salary. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Delihas N. Discovery and characterization of the first non-coding RNA that regulates gene expression, micF RNA: A historical perspective. World J Biol Chem 2015;6:272-80. [Crossref] [PubMed]
- Dinger ME, Pang KC, Mercer TR, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008;4:e1000176. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Anglicheau D, Muthukumar T, Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation 2010;90:105-12. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000;408:86-9. [Crossref] [PubMed]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155-62. [Crossref] [PubMed]
- Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009;11:228-34. [Crossref] [PubMed]
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019;20:21-37. [Crossref] [PubMed]
- O'Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Sood P, Krek A, Zavolan M, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A 2006;103:2746-51. [Crossref] [PubMed]
- Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012;22:125-32. [Crossref] [PubMed]
- Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018;75:193-208. [Crossref] [PubMed]
- Wu D, Yan J, Shen X, et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat Commun 2019;10:3854. [Crossref] [PubMed]
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2:282. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 2016;15:978-87. [Crossref] [PubMed]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 2016;5:19276. [Crossref] [PubMed]
- Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010;38:7248-59. [Crossref] [PubMed]
- Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008;319:1244-7. [Crossref] [PubMed]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013;4:2980. [Crossref] [PubMed]
- Garcia-Martin R, Wang G, Brandão BB, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022;601:446-51. [Crossref] [PubMed]
- Akers JC, Gonda D, Kim R, et al. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 2013;113:1-11. [Crossref] [PubMed]
- Collino F, Deregibus MC, Bruno S, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One 2010;5:e11803. [Crossref] [PubMed]
- Lv Z, Wei Y, Wang D, et al. Argonaute 2 in cell-secreted microvesicles guides the function of secreted miRNAs in recipient cells. PLoS One 2014;9:e103599. [Crossref] [PubMed]
- Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2009;2:ra81. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Djudjaj S, Boor P. Cellular and molecular mechanisms of kidney fibrosis. Mol Aspects Med 2019;65:16-36. [Crossref] [PubMed]
- Kuppe C, Ibrahim MM, Kranz J, et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 2021;589:281-6. [Crossref] [PubMed]
- Fierro-Fernández M, Miguel V, Lamas S. Role of redoximiRs in fibrogenesis. Redox Biol 2016;7:58-67. [Crossref] [PubMed]
- Meng XM, Tang PM, Li J, et al. TGF-β/Smad signaling in renal fibrosis. Front Physiol 2015;6:82. [Crossref] [PubMed]
- Eble JA, de Rezende FF. Redox-relevant aspects of the extracellular matrix and its cellular contacts via integrins. Antioxid Redox Signal 2014;20:1977-93. [Crossref] [PubMed]
- Herrera J, Beisang DJ, Peterson M, et al. Dicer1 Deficiency in the Idiopathic Pulmonary Fibrosis Fibroblastic Focus Promotes Fibrosis by Suppressing MicroRNA Biogenesis. Am J Respir Crit Care Med 2018;198:486-96. [Crossref] [PubMed]
- Davis BN, Hilyard AC, Nguyen PH, et al. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 2010;39:373-84. [Crossref] [PubMed]
- Gomez IG, Nakagawa N, Duffield JS. MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis. Am J Physiol Renal Physiol 2016;310:F931-44. [Crossref] [PubMed]
- Connor KL, Teenan O, Cairns C, et al. Identifying cell-enriched miRNAs in kidney injury and repair. JCI Insight 2020;5:140399. [Crossref] [PubMed]
- Chau BN, Xin C, Hartner J, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 2012;4:121ra18. [Crossref] [PubMed]
- McClelland AD, Herman-Edelstein M, Komers R, et al. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin Sci (Lond) 2015;129:1237-49. [Crossref] [PubMed]
- Wang J, Gao Y, Ma M, et al. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem Biophys 2013;67:537-46. [Crossref] [PubMed]
- Li R, Chung AC, Dong Y, et al. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int 2013;84:1129-44. [Crossref] [PubMed]
- Rogler CE, Levoci L, Ader T, et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology 2009;50:575-84. [Crossref] [PubMed]
- Meng J, Li L, Zhao Y, et al. MicroRNA-196a/b Mitigate Renal Fibrosis by Targeting TGF-β Receptor 2. J Am Soc Nephrol. 2016;27:3006-21. [Crossref] [PubMed]
- Guo R, Hao G, Bao Y, et al. MiR-200a negatively regulates TGF-β1-induced epithelial-mesenchymal transition of peritoneal mesothelial cells by targeting ZEB1/2 expression. Am J Physiol Renal Physiol 2018;314:F1087-F1095. [Crossref] [PubMed]
- Wang B, Koh P, Winbanks C, et al. miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes 2011;60:280-7. [Crossref] [PubMed]
- Gong Y, Qin Z, Zhou B, et al. MicroRNA-200a Inhibits Transforming Growth Factor β1-Induced Proximal Tubular Epithelial-Mesenchymal Transition by Targeting β-Catenin. Nephron 2017;137:237-49. [Crossref] [PubMed]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576-9. [Crossref] [PubMed]
- Wang B, Jha JC, Hagiwara S, et al. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int 2014;85:352-61. [Crossref] [PubMed]
- Liu M, Liu L, Bai M, et al. Hypoxia-induced activation of Twist/miR-214/E-cadherin axis promotes renal tubular epithelial cell mesenchymal transition and renal fibrosis. Biochem Biophys Res Commun 2018;495:2324-30. [Crossref] [PubMed]
- Bai M, Chen H, Ding D, et al. MicroRNA-214 promotes chronic kidney disease by disrupting mitochondrial oxidative phosphorylation. Kidney Int 2019;95:1389-404. [Crossref] [PubMed]
- Hou X, Tian J, Geng J, et al. MicroRNA-27a promotes renal tubulointerstitial fibrosis via suppressing PPARγ pathway in diabetic nephropathy. Oncotarget. 2016;7:47760-76. [Crossref] [PubMed]
- Miguel V, Ramos R, García-Bermejo L, et al. The program of renal fibrogenesis is controlled by microRNAs regulating oxidative metabolism. Redox Biol 2021;40:101851. [Crossref] [PubMed]
- Liu L, Duan A, Guo Q, et al. Detection of microRNA-33a-5p in serum, urine and renal tissue of patients with IgA nephropathy. Exp Ther Med 2021;21:205. [Crossref] [PubMed]
- Wang B, Komers R, Carew R, et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 2012;23:252-65. [Crossref] [PubMed]
- Bijkerk R, de Bruin RG, van Solingen C, et al. Silencing of microRNA-132 reduces renal fibrosis by selectively inhibiting myofibroblast proliferation. Kidney Int 2016;89:1268-80. [Crossref] [PubMed]
- Mann C, Kaistha BP, Kacik M, et al. Downregulation of miR-503 in Activated Kidney Fibroblasts Disinhibits KCNN4 in an in Vitro Model of Kidney Fibrosis. Kidney Blood Press Res 2019;44:113-22. [Crossref] [PubMed]
- Zhou J, Wang KC, Wu W, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci U S A 2011;108:10355-60. [Crossref] [PubMed]
- Song N, Zhang T, Xu X, et al. miR-21 Protects Against Ischemia/Reperfusion-Induced Acute Kidney Injury by Preventing Epithelial Cell Apoptosis and Inhibiting Dendritic Cell Maturation. Front Physiol 2018;9:790. [Crossref] [PubMed]
- Lan YF, Chen HH, Lai PF, et al. MicroRNA-494 reduces ATF3 expression and promotes AKI. J Am Soc Nephrol 2012;23:2012-23. [Crossref] [PubMed]
- Lorenzen JM, Kaucsar T, Schauerte C, et al. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol 2014;25:2717-29. [Crossref] [PubMed]
- Shen Y, Zhao Y, Wang L, et al. MicroRNA-194 overexpression protects against hypoxia/reperfusion-induced HK-2 cell injury through direct targeting Rheb. J Cell Biochem 2018; Epub ahead of print. [Crossref] [PubMed]
- Zhu HY, Liu MY, Hong Q, et al. Role of microRNA-181a in the apoptosis of tubular epithelial cell induced by cisplatin. Chin Med J (Engl) 2012;125:523-6. [PubMed]
- Wei Q, Liu Y, Liu P, et al. MicroRNA-489 Induction by Hypoxia-Inducible Factor-1 Protects against Ischemic Kidney Injury. J Am Soc Nephrol 2016;27:2784-96. [Crossref] [PubMed]
- Ranganathan P, Jayakumar C, Tang Y, et al. MicroRNA-150 deletion in mice protects kidney from myocardial infarction-induced acute kidney injury. Am J Physiol Renal Physiol 2015;309:F551-8. [Crossref] [PubMed]
- Chen HH, Lan YF, Li HF, et al. Urinary miR-16 transactivated by C/EBPβ reduces kidney function after ischemia/reperfusion-induced injury. Sci Rep 2016;6:27945. [Crossref] [PubMed]
- Wang S, Zhang Z, Wang J, et al. MiR-107 induces TNF-α secretion in endothelial cells causing tubular cell injury in patients with septic acute kidney injury. Biochem Biophys Res Commun 2017;483:45-51. [Crossref] [PubMed]
- Abdul-Maksoud RS, Rashad NM, Elsayed WSH, et al. Circulating miR-181a and miR-223 expression with the potential value of biomarkers for the diagnosis of systemic lupus erythematosus and predicting lupus nephritis. J Gene Med 2021;23:e3326. [Crossref] [PubMed]
- Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 2007;104:3432-7. [Crossref] [PubMed]
- Park JT, Kato M, Yuan H, et al. FOG2 protein down-regulation by transforming growth factor-β1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem 2013;288:22469-80. [Crossref] [PubMed]
- Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881-9. [Crossref] [PubMed]
- Zhong X, Chung AC, Chen HY, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013;56:663-74. [Crossref] [PubMed]
- Xu H, Sun F, Li X, et al. Down-regulation of miR-23a inhibits high glucose-induced EMT and renal fibrogenesis by up-regulation of SnoN. Hum Cell 2018;31:22-32. [Crossref] [PubMed]
- Lin CL, Lee PH, Hsu YC, et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol 2014;25:1698-709. [Crossref] [PubMed]
- Long J, Wang Y, Wang W, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 2011;286:11837-48. [Crossref] [PubMed]
- Fu Y, Zhang Y, Wang Z, et al. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am J Nephrol 2010;32:581-9. [Crossref] [PubMed]
- Zhao Y, Yin Z, Li H, et al. MiR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice. Aging Cell 2017;16:387-400. [Crossref] [PubMed]
- Zhao D, Jia J, Shao H. miR-30e targets GLIPR-2 to modulate diabetic nephropathy: in vitro and in vivo experiments. J Mol Endocrinol 2017;59:181-90. [Crossref] [PubMed]
- Bai X, Geng J, Zhou Z, et al. MicroRNA-130b improves renal tubulointerstitial fibrosis via repression of Snail-induced epithelial-mesenchymal transition in diabetic nephropathy. Sci Rep 2016;6:20475. [Crossref] [PubMed]
- Ding H, Li J, Li Y, et al. MicroRNA-10 negatively regulates inflammation in diabetic kidney via targeting activation of the NLRP3 inflammasome. Mol Ther 2021;29:2308-20. [Crossref] [PubMed]
- Wei H, Li J, Li Y, et al. MicroRNA-451 inhibits inflammation and proliferation of glomerular mesangial cells through down-regulating PSMD11 and NF-κB p65. Biosci Rep 2019;39:BSR20191455. [Crossref] [PubMed]
- Sun Y, Peng R, Peng H, et al. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol Cell Endocrinol 2016;433:75-86. [Crossref] [PubMed]
- Wang Q, Wang Y, Minto AW, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 2008;22:4126-35. [Crossref] [PubMed]
- Bera A, Das F, Ghosh-Choudhury N, et al. Reciprocal regulation of miR-214 and PTEN by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. Am J Physiol Cell Physiol 2017;313:C430-47. [Crossref] [PubMed]
- Ma Z, Li L, Livingston MJ, et al. p53/microRNA-214/ULK1 axis impairs renal tubular autophagy in diabetic kidney disease. J Clin Invest 2020;130:5011-26. [Crossref] [PubMed]
- Dong W, Zhang H, Zhao C, et al. Silencing of miR-150-5p Ameliorates Diabetic Nephropathy by Targeting SIRT1/p53/AMPK Pathway. Front Physiol 2021;12:624989. [Crossref] [PubMed]
- Liu Y, Taylor NE, Lu L, et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 2010;55:974-82. [Crossref] [PubMed]
- Cheng Y, Wang D, Wang F, et al. Endogenous miR-204 Protects the Kidney against Chronic Injury in Hypertension and Diabetes. J Am Soc Nephrol 2020;31:1539-54. [Crossref] [PubMed]
- Sun L, Zhang D, Liu F, et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J Pathol 2011;225:364-77. [Crossref] [PubMed]
- Zheng L, Xu CC, Chen WD, et al. MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem Biophys Res Commun 2010;400:483-8. [Crossref] [PubMed]
- Lyu H, Li X, Wu Q, et al. Overexpression of microRNA-21 mediates Ang II-induced renal fibrosis by activating the TGF-β1/Smad3 pathway via suppressing PPARα. J Pharmacol Sci 2019;141:70-8. [Crossref] [PubMed]
- Lu Q, Ma Z, Ding Y, et al. Circulating miR-103a-3p contributes to angiotensin II-induced renal inflammation and fibrosis via a SNRK/NF-κB/p65 regulatory axis. Nat Commun 2019;10:2145. [Crossref] [PubMed]
- Wang Z, Zhu Q, Wang W, et al. Downregulation of microRNA-429 contributes to angiotensin II-induced profibrotic effect in rat kidney. Am J Physiol Renal Physiol 2018;315:F1536-41. [Crossref] [PubMed]
- Serino G, Sallustio F, Cox SN, et al. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol 2012;23:814-24. [Crossref] [PubMed]
- Serino G, Sallustio F, Curci C, et al. Role of let-7b in the regulation of N-acetylgalactosaminyltransferase 2 in IgA nephropathy. Nephrol Dial Transplant 2015;30:1132-9. [Crossref] [PubMed]
- Sun Q, Lan J, Zhang H, et al. MicroRNA-196b targets COSMC in pediatric IgA nephropathy. Mol Med Rep 2020;21:2260-6. [Crossref] [PubMed]
- Bao H, Chen H, Zhu X, et al. MiR-223 downregulation promotes glomerular endothelial cell activation by upregulating importin α4 and α5 in IgA nephropathy. Kidney Int 2014;85:624-35. [Crossref] [PubMed]
- Liang Y, Zhao G, Tang L, et al. MiR-100-3p and miR-877-3p regulate overproduction of IL-8 and IL-1β in mesangial cells activated by secretory IgA from IgA nephropathy patients. Exp Cell Res 2016;347:312-21. [Crossref] [PubMed]
- Guo Y, Liao Y. miR-200bc/429 cluster alleviates inflammation in IgA nephropathy by targeting TWEAK/Fn14. Int Immunopharmacol 2017;52:150-5. [Crossref] [PubMed]
- Bao H, Hu S, Zhang C, et al. Inhibition of miRNA-21 prevents fibrogenic activation in podocytes and tubular cells in IgA nephropathy. Biochem Biophys Res Commun 2014;444:455-60. [Crossref] [PubMed]
- Zhu Y, Xue Z, Di L. Regulation of MiR-146a and TRAF6 in the Diagnose of Lupus Nephritis. Med Sci Monit 2017;23:2550-7. [Crossref] [PubMed]
- Luan J, Fu J, Chen C, et al. LNA-anti-miR-150 ameliorated kidney injury of lupus nephritis by inhibiting renal fibrosis and macrophage infiltration. Arthritis Res Ther 2019;21:276. [Crossref] [PubMed]
- Krasoudaki E, Banos A, Stagakis E, et al. Micro-RNA analysis of renal biopsies in human lupus nephritis demonstrates up-regulated miR-422a driving reduction of kallikrein-related peptidase 4. Nephrol Dial Transplant 2016;31:1676-86. [Crossref] [PubMed]
- Tangtanatakul P, Thammasate B, Jacquet A, et al. Transcriptomic profiling in human mesangial cells using patient-derived lupus autoantibodies identified miR-10a as a potential regulator of IL8. Sci Rep 2017;7:14517. [Crossref] [PubMed]
- Patel V, Williams D, Hajarnis S, et al. miR-17~92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci U S A 2013;110:10765-70. [Crossref] [PubMed]
- Hajarnis S, Lakhia R, Yheskel M, et al. microRNA-17 family promotes polycystic kidney disease progression through modulation of mitochondrial metabolism. Nat Commun 2017;8:14395. [Crossref] [PubMed]
- Shin Y, Kim DY, Ko JY, et al. Regulation of KLF12 by microRNA-20b and microRNA-106a in cystogenesis. FASEB J 2018;32:3574-82. [Crossref] [PubMed]
- Duan J, Huang H, Lv X, et al. PKHD1 post-transcriptionally modulated by miR-365-1 inhibits cell-cell adhesion. Cell Biochem Funct 2012;30:382-9. [Crossref] [PubMed]
- Kim DY, Woo YM, Lee S, et al. Impact of miR-192 and miR-194 on cyst enlargement through EMT in autosomal dominant polycystic kidney disease. FASEB J 2019;33:2870-84. [Crossref] [PubMed]
- Lakhia R, Hajarnis S, Williams D, et al. MicroRNA-21 Aggravates Cyst Growth in a Model of Polycystic Kidney Disease. J Am Soc Nephrol 2016;27:2319-30. [Crossref] [PubMed]
- Sun L, Zhu J, Wu M, et al. Inhibition of MiR-199a-5p reduced cell proliferation in autosomal dominant polycystic kidney disease through targeting CDKN1C. Med Sci Monit 2015;21:195-200. [Crossref] [PubMed]
- Lakhia R, Yheskel M, Flaten A, et al. Interstitial microRNA miR-214 attenuates inflammation and polycystic kidney disease progression. JCI Insight 2020;5:133785. [Crossref] [PubMed]
- Sweetwyne MT, Tao J, Susztak K. Kick it up a notch: Notch signaling and kidney fibrosis. Kidney Int Suppl (2011) 2014;4:91-6. [Crossref] [PubMed]
- Kang HM, Ahn SH, Choi P, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 2015;21:37-46. [Crossref] [PubMed]
- Price NL, Miguel V, Ding W, et al. Genetic deficiency or pharmacological inhibition of miR-33 protects from kidney fibrosis. JCI Insight 2019;4:131102. [Crossref] [PubMed]
- Karpman D, Ståhl AL, Arvidsson I. Extracellular vesicles in renal disease. Nat Rev Nephrol 2017;13:545-62. [Crossref] [PubMed]
- Qi R, Yang C. Renal tubular epithelial cells: the neglected mediator of tubulointerstitial fibrosis after injury. Cell Death Dis 2018;9:1126. [Crossref] [PubMed]
- Borges FT, Melo SA, Özdemir BC, et al. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol 2013;24:385-92. [Crossref] [PubMed]
- Guan H, Peng R, Mao L, et al. Injured tubular epithelial cells activate fibroblasts to promote kidney fibrosis through miR-150-containing exosomes. Exp Cell Res 2020;392:112007. [Crossref] [PubMed]
- Zhou Y, Xiong M, Fang L, et al. miR-21-containing microvesicles from injured tubular epithelial cells promote tubular phenotype transition by targeting PTEN protein. Am J Pathol 2013;183:1183-96. [Crossref] [PubMed]
- Liu JR, Cai GY, Ning YC, et al. Caloric restriction alleviates aging-related fibrosis of kidney through downregulation of miR-21 in extracellular vesicles. Aging (Albany NY) 2020;12:18052-72. [Crossref] [PubMed]
- Zheng SB, Zheng Y, Jin LW, et al. Microvesicles containing microRNA-21 secreted by proximal tubular epithelial cells are involved in renal interstitial fibrosis by activating AKT pathway. Eur Rev Med Pharmacol Sci 2018;22:707-14. [PubMed]
- Qu NY, Zhang ZH, Zhang XX, et al. Microvesicles containing microRNA-216a secreted by tubular epithelial cells participate in renal interstitial fibrosis through activating PTEN/AKT pathway. Eur Rev Med Pharmacol Sci 2019;23:6629-36. [PubMed]
- Zhou Y, Xiong M, Niu J, et al. Secreted fibroblast-derived miR-34a induces tubular cell apoptosis in fibrotic kidney. J Cell Sci 2014;127:4494-506. [PubMed]
- Muralidharan J, Ramezani A, Hubal M, et al. Extracellular microRNA signature in chronic kidney disease. Am J Physiol Renal Physiol 2017;312:F982-91. [Crossref] [PubMed]
- Lange T, Artelt N, Kindt F, et al. MiR-21 is up-regulated in urinary exosomes of chronic kidney disease patients and after glomerular injury. J Cell Mol Med 2019;23:4839-43. [Crossref] [PubMed]
- Glowacki F, Savary G, Gnemmi V, et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One 2013;8:e58014. [Crossref] [PubMed]
- Donderski R, Szczepanek J, Naruszewicz N, et al. Analysis of profibrogenic microRNAs (miRNAs) expression in urine and serum of chronic kidney disease (CKD) stage 1-4 patients and their relationship with proteinuria and kidney function. Int Urol Nephrol 2022;54:937-47. [Crossref] [PubMed]
- Fourdinier O, Schepers E, Metzinger-Le Meuth V, et al. Serum levels of miR-126 and miR-223 and outcomes in chronic kidney disease patients. Sci Rep 2019;9:4477. [Crossref] [PubMed]
- Liu X, Wang W, Bai Y, et al. Identification of a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for chronic kidney disease using next-generation sequencing. J Int Med Res 2020;48:300060520969481. [Crossref] [PubMed]
- Khurana R, Ranches G, Schafferer S, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA 2017;23:142-52. [Crossref] [PubMed]
- Ichii O, Otsuka S, Sasaki N, et al. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int 2012;81:280-92. [Crossref] [PubMed]
- Kellum JA, Romagnani P, Ashuntantang G, et al. Acute kidney injury. Nat Rev Dis Primers 2021;7:52. [Crossref] [PubMed]
- Wei Q, Bhatt K, He HZ, et al. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 2010;21:756-61. [Crossref] [PubMed]
- Xu X, Kriegel AJ, Liu Y, et al. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int 2012;82:1167-75. [Crossref] [PubMed]
- Muratsu-Ikeda S, Nangaku M, Ikeda Y, et al. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS One 2012;7:e41462. [Crossref] [PubMed]
- Aguado-Fraile E, Ramos E, Sáenz-Morales D, et al. miR-127 protects proximal tubule cells against ischemia/reperfusion: identification of kinesin family member 3B as miR-127 target. PLoS One 2012;7:e44305. [Crossref] [PubMed]
- Xu X, Kriegel AJ, Jiao X, et al. miR-21 in ischemia/reperfusion injury: a double-edged sword? Physiol Genomics 2014;46:789-97. [Crossref] [PubMed]
- Lv LL, Wang C, Li ZL, et al. SAP130 released by damaged tubule drives necroinflammation via miRNA-219c/Mincle signaling in acute kidney injury. Cell Death Dis 2021;12:866. [Crossref] [PubMed]
- Li ZL, Lv LL, Tang TT, et al. HIF-1α inducing exosomal microRNA-23a expression mediates the cross-talk between tubular epithelial cells and macrophages in tubulointerstitial inflammation. Kidney Int 2019;95:388-404. [Crossref] [PubMed]
- Lv LL, Feng Y, Wu M, et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death Differ 2020;27:210-26. [Crossref] [PubMed]
- Yu W, Zeng H, Chen J, et al. miR-20a-5p is enriched in hypoxia-derived tubular exosomes and protects against acute tubular injury. Clin Sci (Lond) 2020;134:2223-34. [Crossref] [PubMed]
- Wu XQ, Tian XY, Wang ZW, et al. miR-191 secreted by platelet-derived microvesicles induced apoptosis of renal tubular epithelial cells and participated in renal ischemia-reperfusion injury via inhibiting CBS. Cell Cycle 2019;18:119-29. [Crossref] [PubMed]
- Brandenburger T, Lorenzen JM. Diagnostic and Therapeutic Potential of microRNAs in Acute Kidney Injury. Front Pharmacol 2020;11:657. [Crossref] [PubMed]
- Du J, Cao X, Zou L, et al. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS One 2013;8:e63390. [Crossref] [PubMed]
- Ramachandran K, Saikumar J, Bijol V, et al. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem 2013;59:1742-52. [Crossref] [PubMed]
- Wang N, Zhou Y, Jiang L, et al. Urinary microRNA-10a and microRNA-30d serve as novel, sensitive and specific biomarkers for kidney injury. PLoS One 2012;7:e51140. [Crossref] [PubMed]
- Sonoda H, Lee BR, Park KH, et al. miRNA profiling of urinary exosomes to assess the progression of acute kidney injury. Sci Rep 2019;9:4692. [Crossref] [PubMed]
- Aguado-Fraile E, Ramos E, Conde E, et al. A Pilot Study Identifying a Set of microRNAs As Precise Diagnostic Biomarkers of Acute Kidney Injury. PLoS One 2015;10:e0127175. [Crossref] [PubMed]
- Lorenzen JM, Kielstein JT, Hafer C, et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol 2011;6:1540-6. [Crossref] [PubMed]
- Pavkovic M, Riefke B, Ellinger-Ziegelbauer H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 2014;324:147-57. [Crossref] [PubMed]
- Sun SQ, Zhang T, Ding D, et al. Circulating MicroRNA-188, -30a, and -30e as Early Biomarkers for Contrast-Induced Acute Kidney Injury. J Am Heart Assoc 2016;5:004138. [Crossref] [PubMed]
- Liu Z, Tang C, He L, et al. The negative feedback loop of NF-κB/miR-376b/NFKBIZ in septic acute kidney injury. JCI Insight 2020;5:e142272. [Crossref]
- Fan PC, Chen CC, Peng CC, et al. A circulating miRNA signature for early diagnosis of acute kidney injury following acute myocardial infarction. J Transl Med 2019;17:139. [Crossref] [PubMed]
- Thomas MC, Brownlee M, Susztak K, et al. Diabetic kidney disease. Nat Rev Dis Primers 2015;1:15018. [Crossref] [PubMed]
- Harvey SJ, Jarad G, Cunningham J, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 2008;19:2150-8. [Crossref] [PubMed]
- Zhdanova O, Srivastava S, Di L, et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int 2011;80:719-30. [Crossref] [PubMed]
- Chung AC, Huang XR, Meng X, et al. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 2010;21:1317-25. [Crossref] [PubMed]
- Hu R, Li X, Peng C, et al. miR-196b-5p-enriched extracellular vesicles from tubular epithelial cells mediated aldosterone-induced renal fibrosis in mice with diabetes. BMJ Open Diabetes Res Care 2020;8:e001101. [Crossref] [PubMed]
- Jia Y, Zheng Z, Xue M, et al. Extracellular Vesicles from Albumin-Induced Tubular Epithelial Cells Promote the M1 Macrophage Phenotype by Targeting Klotho. Mol Ther 2019;27:1452-66. [Crossref] [PubMed]
- Jia Y, Zheng Z, Yang Y, et al. MiR-4756 promotes albumin-induced renal tubular epithelial cell epithelial-to-mesenchymal transition and endoplasmic reticulum stress via targeting Sestrin2. J Cell Physiol 2019;234:2905-15. [Crossref] [PubMed]
- Jia Y, Zheng Z, Guan M, et al. Exendin-4 ameliorates high glucose-induced fibrosis by inhibiting the secretion of miR-192 from injured renal tubular epithelial cells. Exp Mol Med 2018;50:1-13. [Crossref] [PubMed]
- Jeon JS, Kim E, Bae YU, et al. microRNA in Extracellular Vesicles Released by Damaged Podocytes Promote Apoptosis of Renal Tubular Epithelial Cells. Cells 2020;9:1409. [Crossref] [PubMed]
- Ghai V, Wu X, Bheda-Malge A, et al. Genome-wide Profiling of Urinary Extracellular Vesicle microRNAs Associated With Diabetic Nephropathy in Type 1 Diabetes. Kidney Int Rep 2018;3:555-72. [Crossref] [PubMed]
- Barutta F, Tricarico M, Corbelli A, et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One 2013;8:e73798. [Crossref] [PubMed]
- Ma X, Lu C, Lv C, et al. The Expression of miR-192 and Its Significance in Diabetic Nephropathy Patients with Different Urine Albumin Creatinine Ratio. J Diabetes Res 2016;2016:6789402. [Crossref] [PubMed]
- Jia Y, Guan M, Zheng Z, et al. miRNAs in Urine Extracellular Vesicles as Predictors of Early-Stage Diabetic Nephropathy. J Diabetes Res 2016;2016:7932765. [Crossref] [PubMed]
- Wang J, Duan L, Tian L, et al. Serum miR-21 may be a Potential Diagnostic Biomarker for Diabetic Nephropathy. Exp Clin Endocrinol Diabetes 2016;124:417-23. [PubMed]
- Zang J, Maxwell AP, Simpson DA, et al. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci Rep 2019;9:10900. [Crossref] [PubMed]
- Mohan A, Singh RS, Kumari M, et al. Urinary Exosomal microRNA-451-5p Is a Potential Early Biomarker of Diabetic Nephropathy in Rats. PLoS One 2016;11:e0154055. [Crossref] [PubMed]
- Park S, Moon S, Lee K, et al. Urinary and Blood MicroRNA-126 and -770 are Potential Noninvasive Biomarker Candidates for Diabetic Nephropathy: a Meta-Analysis. Cell Physiol Biochem 2018;46:1331-40. [Crossref] [PubMed]
- Shao Y, Ren H, Lv C, et al. Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine 2017;55:130-8. [Crossref] [PubMed]
- Lv C, Zhou YH, Wu C, et al. The changes in miR-130b levels in human serum and the correlation with the severity of diabetic nephropathy. Diabetes Metab Res Rev 2015;31:717-24. [Crossref] [PubMed]
- Han Q, Zhang Y, Jiao T, et al. Urinary sediment microRNAs can be used as potential noninvasive biomarkers for diagnosis, reflecting the severity and prognosis of diabetic nephropathy. Nutr Diabetes 2021;11:24. [Crossref] [PubMed]
- Yang X, Wu D, Du H, et al. MicroRNA-135a is involved in podocyte injury in a transient receptor potential channel 1-dependent manner. Int J Mol Med 2017;40:1511-9. [Crossref] [PubMed]
- Delić D, Eisele C, Schmid R, et al. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS One 2016;11:e0150154. [Crossref] [PubMed]
- Eissa S, Matboli M, Aboushahba R, et al. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J Diabetes Complications 2016;30:1585-92. [Crossref] [PubMed]
- Zhang L, He S, Guo S, et al. Down-regulation of miR-34a alleviates mesangial proliferation in vitro and glomerular hypertrophy in early diabetic nephropathy mice by targeting GAS1. J Diabetes Complications 2014;28:259-64. [Crossref] [PubMed]
- Yan JD, Yang S, Zhang J, et al. BMP6 reverses TGF-beta1-induced changes in HK-2 cells: implications for the treatment of renal fibrosis. Acta Pharmacol Sin 2009;30:994-1000. [Crossref] [PubMed]
- Zhao Y, Shen A, Guo F, et al. Urinary Exosomal MiRNA-4534 as a Novel Diagnostic Biomarker for Diabetic Kidney Disease. Front Endocrinol (Lausanne) 2020;11:590. [Crossref] [PubMed]
- Lin M, Song D, Zhang S, et al. Dysregulation of miR-638 in diabetic nephropathy and its role in inflammatory response. Diabetol Metab Syndr 2021;13:122. [Crossref] [PubMed]
- Agarwal R. Hypertension in chronic kidney disease and dialysis: pathophysiology and management. Cardiol Clin 2005;23:237-48. [Crossref] [PubMed]
- Wang G, Kwan BC, Lai FM, et al. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens 2010;23:78-84. [Crossref] [PubMed]
- Chen C, Lu C, Qian Y, et al. Urinary miR-21 as a potential biomarker of hypertensive kidney injury and fibrosis. Sci Rep 2017;7:17737. [Crossref] [PubMed]
- Mennuni S, Rubattu S, Pierelli G, et al. Hypertension and kidneys: unraveling complex molecular mechanisms underlying hypertensive renal damage. J Hum Hypertens 2014;28:74-9. [Crossref] [PubMed]
- Gildea JJ, Carlson JM, Schoeffel CD, et al. Urinary exosome miRNome analysis and its applications to salt sensitivity of blood pressure. Clin Biochem 2013;46:1131-4. [Crossref] [PubMed]
- Zununi Vahed S, Poursadegh Zonouzi A, Ghanbarian H, et al. Differential expression of circulating miR-21, miR-142-3p and miR-155 in renal transplant recipients with impaired graft function. Int Urol Nephrol 2017;49:1681-9. [Crossref] [PubMed]
- Atala A. Re: Circulating miR-103a-3p Contributes to Angiotensin II-Induced Renal Inflammation and Fibrosis via a SNRK/NF-κB/p65 Regulatory Axis. J Urol 2020;203:34-5. [Crossref] [PubMed]
- Perez-Hernandez J, Olivares D, Forner MJ, et al. Urinary exosome miR-146a is a potential marker of albuminuria in essential hypertension. J Transl Med 2018;16:228. [Crossref] [PubMed]
- Huang YQ, Huang C, Li J, et al. The association of miR-29a with proteinuria in essential hypertension. J Hum Hypertens 2018;32:775-80. [Crossref] [PubMed]
- Perez-Hernandez J, Riffo-Campos AL, Ortega A, et al. Urinary- and Plasma-Derived Exosomes Reveal a Distinct MicroRNA Signature Associated With Albuminuria in Hypertension. Hypertension 2021;77:960-71. [Crossref] [PubMed]
- Suzuki H, Kiryluk K, Novak J, et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol 2011;22:1795-803. [Crossref] [PubMed]
- Lech M, Anders HJ. The pathogenesis of lupus nephritis. J Am Soc Nephrol 2013;24:1357-66. [Crossref] [PubMed]
- Wang G, Kwan BC, Lai FM, et al. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest 2010;90:98-103. [Crossref] [PubMed]
- Wang G, Kwan BC, Lai FM, et al. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers 2011;30:171-9. [Crossref] [PubMed]
- Pawluczyk IZA, Didangelos A, Barbour SJ, et al. Differential expression of microRNA miR-150-5p in IgA nephropathy as a potential mediator and marker of disease progression. Kidney Int 2021;99:1127-39. [Crossref] [PubMed]
- Serino G, Pesce F, Sallustio F, et al. In a retrospective international study, circulating miR-148b and let-7b were found to be serum markers for detecting primary IgA nephropathy. Kidney Int 2016;89:683-92. [Crossref] [PubMed]
- Wu J, Zhang H, Wang W, et al. Plasma microRNA signature of patients with IgA nephropathy. Gene 2018;649:80-6. [Crossref] [PubMed]
- Fan Q, Lu R, Zhu M, et al. Serum miR-192 Is Related to Tubulointerstitial Lesion and Short-Term Disease Progression in IgA Nephropathy. Nephron 2019;142:195-207. [Crossref] [PubMed]
- Wang G, Kwan BC, Lai FM, et al. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol 2012;36:412-8. [Crossref] [PubMed]
- Szeto CC, Wang G, Ng JK, et al. Urinary miRNA profile for the diagnosis of IgA nephropathy. BMC Nephrol 2019;20:77. [Crossref] [PubMed]
- Min QH, Chen XM, Zou YQ, et al. Differential expression of urinary exosomal microRNAs in IgA nephropathy. J Clin Lab Anal 2018; [Crossref] [PubMed]
- Duan ZY, Cai GY, Bu R, et al. Selection of urinary sediment miRNAs as specific biomarkers of IgA nephropathy. Sci Rep 2016;6:23498. [Crossref] [PubMed]
- Dai Y, Sui W, Lan H, et al. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int 2009;29:749-54. [Crossref] [PubMed]
- Lu J, Kwan BC, Lai FM, et al. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton) 2012;17:346-51. [Crossref] [PubMed]
- Zhou H, Hasni SA, Perez P, et al. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J Am Soc Nephrol 2013;24:1073-87. [Crossref] [PubMed]
- Guan J, Wang G, Tam LS, et al. Urinary sediment ICAM-1 level in lupus nephritis. Lupus 2012;21:1190-5. [Crossref] [PubMed]
- Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One 2010;5:e10344. [Crossref] [PubMed]
- Carlsen AL, Schetter AJ, Nielsen CT, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum 2013;65:1324-34. [Crossref] [PubMed]
- Perez-Hernandez J, Martinez-Arroyo O, Ortega A, et al. Urinary exosomal miR-146a as a marker of albuminuria, activity changes and disease flares in lupus nephritis. J Nephrol 2021;34:1157-67. [Crossref] [PubMed]
- Costa-Reis P, Russo PA, Zhang Z, et al. The Role of MicroRNAs and Human Epidermal Growth Factor Receptor 2 in Proliferative Lupus Nephritis. Arthritis Rheumatol 2015;67:2415-26. [Crossref] [PubMed]
- Garcia-Vives E, Solé C, Moliné T, et al. The Urinary Exosomal miRNA Expression Profile is Predictive of Clinical Response in Lupus Nephritis. Int J Mol Sci 2020;21:1372. [Crossref] [PubMed]
- Solé C, Cortés-Hernández J, Felip ML, et al. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol Dial Transplant 2015;30:1488-96. [Crossref] [PubMed]
- Solé C, Moliné T, Vidal M, et al. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells 2019;8:773. [Crossref] [PubMed]
- Li Y, Xu X, Tang X, et al. MicroRNA expression profile of urinary exosomes in Type IV lupus nephritis complicated by cellular crescent. J Biol Res (Thessalon) 2018;25:16. [Crossref] [PubMed]
- Igarashi P, Somlo S. Genetics and pathogenesis of polycystic kidney disease. J Am Soc Nephrol 2002;13:2384-98. [Crossref] [PubMed]
- Pandey P, Qin S, Ho J, et al. Systems biology approach to identify transcriptome reprogramming and candidate microRNA targets during the progression of polycystic kidney disease. BMC Syst Biol 2011;5:56. [Crossref] [PubMed]
- Dweep H, Sticht C, Kharkar A, et al. Parallel analysis of mRNA and microRNA microarray profiles to explore functional regulatory patterns in polycystic kidney disease: using PKD/Mhm rat model. PLoS One 2013;8:e53780. [Crossref] [PubMed]
- Tran U, Zakin L, Schweickert A, et al. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development 2010;137:1107-16. [Crossref] [PubMed]
- Ding H, Li LX, Harris PC, et al. Extracellular vesicles and exosomes generated from cystic renal epithelial cells promote cyst growth in autosomal dominant polycystic kidney disease. Nat Commun 2021;12:4548. [Crossref] [PubMed]
- Małachowska B, Tkaczyk M, Chrul S, et al. Serum microRNA profiles in patients with autosomal dominant polycystic kidney disease show systematic dysregulation partially reversible by hemodialysis. Arch Med Sci 2021;17:1730-41. [PubMed]
- Ben-Dov IZ, Tan YC, Morozov P, et al. Urine microRNA as potential biomarkers of autosomal dominant polycystic kidney disease progression: description of miRNA profiles at baseline. PLoS One 2014;9:e86856. [Crossref] [PubMed]
- Kocyigit I, Taheri S, Sener EF, et al. Serum micro-rna profiles in patients with autosomal dominant polycystic kidney disease according to hypertension and renal function. BMC Nephrol 2017;18:179. [Crossref] [PubMed]
- Hill NR, Fatoba ST, Oke JL, et al. Global Prevalence of Chronic Kidney Disease - A Systematic Review and Meta-Analysis. PLoS One 2016;11:e0158765. [Crossref] [PubMed]
- Wonnacott A, Bowen T, Fraser DJ. MicroRNAs as biomarkers in chronic kidney disease. Curr Opin Nephrol Hypertens 2017;26:460-6. [Crossref] [PubMed]
- Gurung S, Perocheau D, Touramanidou L, et al. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal 2021;19:47. [Crossref] [PubMed]
- Jing H, Tang S, Lin S, et al. The role of extracellular vesicles in renal fibrosis. Cell Death Dis 2019;10:367. [Crossref] [PubMed]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010;327:198-201. [Crossref] [PubMed]
- Bose RJ, Lee SH, Park H. Lipid-based surface engineering of PLGA nanoparticles for drug and gene delivery applications. Biomater Res 2016;20:34. [Crossref] [PubMed]
- Endo-Takahashi Y, Negishi Y. Microbubbles and Nanobubbles with Ultrasound for Systemic Gene Delivery. Pharmaceutics 2020;12:964. [Crossref] [PubMed]
- O'Brien K, Breyne K, Ughetto S, et al. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol 2020;21:585-606. [Crossref] [PubMed]
- Zhou Y, Fang L, Yu Y, et al. Erythropoietin protects the tubular basement membrane by promoting the bone marrow to release extracellular vesicles containing tPA-targeting miR-144. Am J Physiol Renal Physiol 2016;310:F27-40. [Crossref] [PubMed]
- Wang B, Yao K, Huuskes BM, et al. Mesenchymal Stem Cells Deliver Exogenous MicroRNA-let7c via Exosomes to Attenuate Renal Fibrosis. Mol Ther 2016;24:1290-301. [Crossref] [PubMed]
- Wang H, Wang B, Zhang A, et al. Exosome-Mediated miR-29 Transfer Reduces Muscle Atrophy and Kidney Fibrosis in Mice. Mol Ther 2019;27:571-83. [Crossref] [PubMed]
- Cantaluppi V, Gatti S, Medica D, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int 2012;82:412-27. [Crossref] [PubMed]
- Tapparo M, Bruno S, Collino F, et al. Renal Regenerative Potential of Extracellular Vesicles Derived from miRNA-Engineered Mesenchymal Stromal Cells. Int J Mol Sci 2019;20:2381. [Crossref] [PubMed]
- Zhang R, Zhu Y, Li Y, et al. Human umbilical cord mesenchymal stem cell exosomes alleviate sepsis-associated acute kidney injury via regulating microRNA-146b expression. Biotechnol Lett 2020;42:669-79. [Crossref] [PubMed]
- Zhu G, Pei L, Lin F, et al. Exosomes from human-bone-marrow-derived mesenchymal stem cells protect against renal ischemia/reperfusion injury via transferring miR-199a-3p. J Cell Physiol 2019;234:23736-49. [Crossref] [PubMed]
- Zhang J, He W, Zheng D, et al. Exosomal-miR-1184 derived from mesenchymal stem cells alleviates cisplatin-associated acute kidney injury. Mol Med Rep 2021;24:795. [Crossref] [PubMed]
- Gu D, Zou X, Ju G, et al. Mesenchymal Stromal Cells Derived Extracellular Vesicles Ameliorate Acute Renal Ischemia Reperfusion Injury by Inhibition of Mitochondrial Fission through miR-30. Stem Cells Int 2016;2016:2093940. [Crossref] [PubMed]
- Viñas JL, Burger D, Zimpelmann J, et al. Transfer of microRNA-486-5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int 2016;90:1238-50. [Crossref] [PubMed]
- Cao H, Cheng Y, Gao H, et al. In Vivo Tracking of Mesenchymal Stem Cell-Derived Extracellular Vesicles Improving Mitochondrial Function in Renal Ischemia-Reperfusion Injury. ACS Nano 2020;14:4014-26. [Crossref] [PubMed]
- Zhong L, Liao G, Wang X, et al. Mesenchymal stem cells-microvesicle-miR-451a ameliorate early diabetic kidney injury by negative regulation of P15 and P19. Exp Biol Med (Maywood) 2018;243:1233-42. [Crossref] [PubMed]
- Mao R, Shen J, Hu X. BMSCs-derived exosomal microRNA-let-7a plays a protective role in diabetic nephropathy via inhibition of USP22 expression. Life Sci 2021;268:118937. [Crossref] [PubMed]
- Duan Y, Luo Q, Wang Y, et al. Adipose mesenchymal stem cell-derived extracellular vesicles containing microRNA-26a-5p target TLR4 and protect against diabetic nephropathy. J Biol Chem 2020;295:12868-84. [Crossref] [PubMed]
- Birtwistle L, Chen XM, Pollock C. Mesenchymal Stem Cell-Derived Extracellular Vesicles to the Rescue of Renal Injury. Int J Mol Sci 2021;22:6596. [Crossref] [PubMed]
Cite this article as: Miguel V. The extracellular miRNA fingerprint of kidney disease: a narrative review. ExRNA 2022;4:12.


