
Extracellular tRNA-derived RNAs as emerging activators of endosomal Toll-like receptors: a narrative review
Introduction
One of the first lines of defense against invading pathogens begins when the pattern-recognition receptors (PRRs) of the innate immune system detect pathogen-associated molecular patterns (PAMPs), leading to initiation of protective responses (1,2). Toll-like receptors (TLRs) are the most extensively studied PRRs, and are expressed in both innate immune cells (e.g., macrophages and dendritic cells) and non-immune cells (e.g., epithelial cells and fibroblast cells) (2-4). Among the 10 TLRs characterized in humans, TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 localize to the cell surface (surface TLRs), while TLR3, TLR7, TLR8, and TLR9 localize to endosomes and other intracellular compartments (endosomal TLRs). Each TLR specifically recognizes a different PAMP, ranging from polysaccharides and lipopeptides to nucleic acids (2-4). Endosomal TLRs are nucleic acid-sensing TLRs, and among those, TLR7 and TLR8 recognize single-stranded RNAs (ssRNAs) as their ligands (2-5). ssRNA recognition by these TLRs results in the recruitment of the adaptor protein MyD88, leading to NF-κB-mediated transcription and downstream induction of interferon and cytokine production (2-4). While “foreign” ssRNAs from bacteria or viruses have been extensively studied as the ligands recognized by TLR7 and TLR8 (5-10), “endogenous” ssRNA ligands from host cells have not been fully characterized yet, and indeed the idea of endogenous ligands of PRRs remains controversial, as it complicates the traditional view of the role of PRRs in distinguishing self from non-self (11).
Once endogenous ssRNA molecules are packaged into extracellular vesicles (EVs), they can be delivered into the endosomes of recipient cells, where they can be sensed by TLR7 or TLR8. This has been observed with certain extracellular (ex-) microRNAs (miRNAs), which, upon delivery to recipient cell endosomes, become ligands for TLR7 and TLR8 and activate the downstream pathway (12,13). This miRNA-mediated TLR7/TLR8 activation is relevant not only to immune response (14,15) but has also been demonstrated to play roles in neurodegeneration (12), tumor growth and metastasis (16,17), autoimmunity (18), and pathobiology of various other diseases (19). Because miRNAs are the best-studied short non-coding RNAs (sncRNAs), it is natural that they have dominated current research on both ex-sncRNAs and endogenous ssRNA ligands of TLRs. However, recent advances in our understanding of “previously-hidden” sncRNAs have widened the pool of candidate ssRNA molecules that could act as endogenous ligands of ssRNA-sensing immune receptors, exemplified in the recent finding that transfer RNA (tRNA)-derived sncRNAs can function as endogenous ligands of TLR7 (20). We present this article in accordance with the Narrative Review reporting checklist (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-22/rc).
Methods
To identify articles suitable for this review, we conducted a literature search via PubMed using search terms that included “tRNA”, “tRNA half”, “tRF”, “tsRNA”, “exRNA”, “circulating RNA”, “immune response”, “TLR”, “TLR7”, “TLR8”, “neurodegeneration”, and “cancer”. Articles written in English and published prior to August 31, 2022 were considered. These details are compiled in Table 1.
Table 1
| Items | Specification |
|---|---|
| Date of search | August 31, 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | tRNA, tRNA half, tRF, tsRNA, exRNA, circulating RNA, immune response, TLR, TLR7, TLR8, neurodegeneration, cancer |
| Timeframe | 1986–2022 |
| Inclusion and exclusion criteria | Research and review manuscripts written in English were included, and articles written in a language other than English were excluded |
| Selection process | Both authors performed the search for suitable articles |
Most ex-sncRNAs are uncaptured by standard RNA-seq
Cellular sncRNA molecules generally possess either a hydroxyl group (OH), a monophosphate (P), or a 2',3'-cyclic phosphate (cP) at their termini (Figure 1A), and the terminal states of each sncRNA are determined by the catalytic machinery underlying the RNA cleavage that produces them (21,24). Although next-generation sequencing of RNA molecules (RNA-seq) has become a common tool to characterize RNA expression profiles, most sncRNA sequencing studies to date have relied on a standard small RNA-seq method in which 5'- and 3'-adaptors (AD) can be ligated only to 5'-P and 3'-OH ends of RNAs, respectively. While this standard method is suitable for efficient amplification and sequencing of 5'-P/3'-OH-containing sncRNAs, such as miRNAs, it cannot capture RNAs with other terminal structures (i.e., 5'-OH, 3'-P, or cP) (Figure 1B). Due to this limitation, non-miRNA-sncRNAs lacking the 5'-P/3'-OH ends have been significantly underrepresented in many of the current sncRNA analyses, and thus comprise a largely unexplored component in the transcriptome.
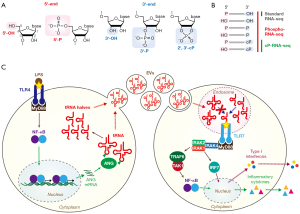
This notion is especially important when it comes to the sequencing analyses of ex-sncRNAs. Human plasma samples contain many ex-sncRNAs which lack 5'-P or 3'-OH and are therefore uncaptured by standard RNA-seq (25). To sequence ex-sncRNAs with any given set of terminal phosphate states, Giraldez et al. developed Phospho-RNA-seq (25) (Figure 1B), which begins with the treatment of RNA samples with T4 polynucleotide kinase (T4 PNK). Because T4 PNK possesses both 5'-phosphorylation (convert 5'-OH to 5'-P in the presence of ATP) and 3'-dephosphorylation (convert 3'-P/cP to 3'-OH) activities, the T4 PNK treatment leaves all RNA species with the 5'-P/3'-OH-ends, thus rendering them available for 5'- and 3'-AD ligation in subsequent small RNA-seq procedure. Phospho-RNA-seq has been successfully used to profile the expression of ex-sncRNAs in human plasma samples and their tissue specific signatures (25). Other human plasma sequencing studies also showed that the addition of T4 PNK treatment to the sequencing procedure significantly altered the proportion of the reads of the sncRNAs derived from tRNAs, messenger RNAs (mRNAs), ribosomal RNAs (rRNAs), and other non-coding RNAs (26,27), corroborating that standard RNA-seq without T4 PNK treatment cannot fully capture these ex-sncRNAs. Our study on EVs secreted from human monocyte-derived macrophages (HMDMs) further confirmed the necessity of T4 PNK treatment in ex-sncRNA sequencing (20). T4 PNK treatment was required to amplify the majority of the cDNAs derived from the EV ex-sncRNAs. Treatment with a mutant T4 PNK, which lacks 3'-dephosphorylation activity, resulted in dramatic reduction of cDNA yield, suggesting that most ex-sncRNAs in HMDM EVs are 3'-P- or cP-containing RNAs, and miRNAs and other 3'-OH containing RNAs comprise only a small part of those ex-sncRNA species packaged into the EVs (20). Based on these findings, recent ex-sncRNA sequencing studies have been utilizing T4 PNK treatment to make terminal formation of RNA molecules consistently 5'-P and 3'-OH, allowing for the capture of the full range of ex-sncRNAs.
tRNA-derived sncRNAs as a component of the expanding sncRNA world
It is well established that tRNAs, canonically understood as adaptor molecules of translational machinery, are also a source of abundant functional sncRNAs (28-31). The tRNA-derived sncRNAs are generated from mature tRNAs or their precursor transcripts, not as random degradation products but as specific functional molecules, and can be classified into two groups: tRNA halves and tRNA-derived fragments (tRFs) (28-31). The 30–35-nt tRNA halves are generally much more abundant than tRFs and are produced by endonucleolytic cleavage of the tRNA anticodon-loop. In mammalian cells, angiogenin (ANG), a member of the RNase A superfamily, is one of the major enzymes responsible for the anticodon cleavage (32,33). Because ANG-mediated cleavage leaves a cP and 5'-P in 5'- and 3'-cleavage products (34), respectively, 5'-tRNA halves contain a 5'-P (from mature tRNAs) and a cP, while 3'-tRNA halves possess a 5'-OH and an amino acid (AA) at the 3'-end (from mature tRNAs), which has been experimentally validated (35). These terminal formations make tRNA half molecules the aforementioned “hidden class” of sncRNAs that are uncaptured by standard RNA-seq. Due to the presence of a cP in 5'-tRNA halves, a specific sequencing method for cP-containing RNAs, termed cP-RNA-seq (Figure 1B), has been utilized for the expressional characterization of 5'-tRNA halves (20,35-39). ANG-mediated tRNA cleavage can be induced by various biological factors/phenomena such as stress stimuli (32,33,38) and sex hormone signaling pathways (35). Although ANG is bound by its inhibitor ribonuclease/angiogenin inhibitor 1 (RNH1) in the cytoplasm, under stress conditions, degradation of RNH1 and/or translocation of ANG to stress granules, where it dissociates from RNH1, enable ANG-mediated tRNA cleavage (40-42). tRNA halves function in stress granule formation (43), regulation of translation (32,44), and promotion of cell proliferation (35); they are also associated with various disorders such as neurodegeneration (45,46). tRNA halves can further serve as direct precursors for shorter sncRNAs such as Piwi-interacting RNAs (piRNAs) (36,39).
tRFs are generally shorter than tRNA halves and can mainly be subclassified into 5'-tRFs, 3'-tRFs, and internal (i)-tRFs (28-31). While 5'- and 3'-tRFs are derived from 5'- and 3'-parts of mature tRNAs, respectively, i-tRFs are derived wholly from internal parts of mature tRNAs. Although Dicer and ANG are known to be involved in the biogenesis of some tRFs (47,48), detailed regulatory mechanisms and other biogenesis factors for tRFs remain elusive, leaving terminal phosphate states of tRFs undefined. This further emphasizes the importance of inclusive sequencing methods such as Phospho-RNA-seq for the comprehensive profiling of tRNA-derived sncRNAs. While many tRFs have been shown to function as miRNAs or piRNAs by binding to AGO or PIWI proteins (29,36,39), tRFs further have various functions, such as regulating mRNA stability or translation, preventing cell apoptosis, and promoting viral infection, and their dysregulation is involved in various diseases (28,30,31,49).
tRNA-derived sncRNAs as abundant ex-RNAs
tRNA-derived sncRNAs are increasingly recognized as an abundant class of ex-RNAs, released from cells under diverse conditions and packaged into EVs (50-52) or bound to RNA-binding proteins (RNPs) (53) or lipoprotein particles (LPPs) (54). Functional roles of some of these tRNA-derived ex-sncRNAs have also been identified. For example, ex-5'-tRNA halves and ex-5'-tRFs contained in the vesicles deriving from the epididymis, termed epididymosomes, are transferred to maturing sperm (55) and regulate gene expressions in embryonic stem cells and embryos (55-57). tRNA derived ex-sncRNAs further appear to have potential as biomarkers in various disease contexts (58). For example, tRNA-derived ex-sncRNAs are differentially accumulated in breast cancer (59), prostate cancer (60), lung cancer (61), and chronic kidney disease (62), compared to healthy individuals. For further discussion of tRNA-derived ex-sncRNAs, we recommend a review from Tosar and Cayota (63). Experimental results on tRNA-derived ex-sncRNAs have continuously accumulated; more recent studies have provided further evidence that tRNA-derived ex-sncRNAs could be useful prognostic and diagnostic biomarkers in breast cancer (64-66), and have also shown their differential accumulation in metastatic hypopharyngeal cancer (67), systemic lupus erythematosus (68), and ischemic kidney injury (69).
Infection-induced ex-tRNA halves activate TLR7
We recently identified a novel role of ex-tRNA halves as activators of endosomal TLR (20). We demonstrated that infection of HMDMs with Mycobacterium bovis BCG and surface TLR activation by treatment with lipopolysaccharide (LPS) or peptidoglycan (PGN) upregulate the NF-κB-mediated transcription of ANG, leading to accumulation of tRNA half molecules in HMDMs and their secreted EVs. In sequencing data of T4 PNK-treated RNAs from the EVs, 5'-tRNA halves comprised over 96% of tRNA-derived reads [in EV #1 library (20)], while 3'-tRNA halves (1.3%), 5'-tRFs (0.87%), 3'-tRFs (0.63%), and i-tRFs (0.99%) were minor species. The mechanisms underlying the specific and selective packaging of tRNA-derived sncRNAs into EVs are unknown, but specific RNA binding proteins, such as YBX1 that binds to 5'-tRNA halves (44) and functions in RNA sorting into EVs (70), could be involved in the mechanisms. Most ex-5'-tRNA halves were derived from a focused subset of cytoplasmic tRNAs, including tRNAValCAC/AAC, tRNAGlyGCC, tRNAHisGUG, and tRNAGluCUC, which in aggregate were the sources of approximately 90% of the identified ex-5'-tRNA halves. One of the most remarkable characteristics of ex-tRNA halves is their abundance. The ex-5'-tRNAHisGUG half was >210 times more abundant than miR-150, one of the most abundant miRNAs in HMDM-derived EVs (20). While it has been demonstrated that ex-miRNAs can act as ligands for endosomal TLRs, the much greater abundance of ex-tRNA halves suggests that they might constitute a more significant, biologically relevant class of endogenous ligands of these immune receptors. Indeed, the EV-contained ex-tRNA halves were experimentally confirmed to be delivered into endosomes in recipient HMDMs where the 5'-tRNAHisGUG half strongly activates TLR7 (20). The activity of the 5'-tRNAHisGUG half is as high as that of HIV-derived ssRNA40, a widely used positive control ssRNA known as a strong activator of endosomal TLRs (5), suggesting that 5'-tRNAHisGUG half could have the capacity to produce an immune response.
Expressional induction and secretion of tRNA halves are not limited to cell culture settings but have been further observed in actual pathological situations. By developing a sensitive multiplex tRNA half quantification method, we revealed an approximately 1,000-fold enrichment of plasma ex-5'-tRNA halves in patients infected with Mycobacterium tuberculosis (20,71). A dramatic increase in the levels of serum ex-5'-tRNA halves has also been observed in LPS-injected mice and monkeys and in patients experiencing active, but not quiescent, hepatitis B virus infection (72). Because upregulation of 5'-tRNA half expression has been reported upon infection with respiratory syncytial virus (73,74), Rickettsia (75), and hepatitis B and C viruses (76), it seems possible that induction of 5'-tRNA halves and their secretion as ex-5'-tRNA halves could be a universal phenomenon among infectious diseases. These studies suggest a novel role of ex-5'-tRNA half molecules as “immune activators,” but further studies are required to fully unveil the immune response pathways mediated by tRNA-derived ex-sncRNAs.
Future perspectives
The study of endogenous ex-sncRNA ligands of immune receptors is still at an initial stage and remains an area of some controversy. Likewise, we are just beginning to appreciate the previously hidden classes of sncRNAs not detected by standard RNA-seq methods. We expect that the 5'-tRNAHisGUG half is just the first example of an immune receptor-simulating tRNA-derived ex-sncRNA, and other tRNA halves and tRFs could be found to be similarly immunostimulatory through the TLR7 or TLR8 axis. The 5'-tRNAHisGUG half strongly activates TLR7, but not TLR8 (20), probably because of differences in ligand specificity for TLR7 and TLR8 (6,77,78). Future exploration of immune stimulatory tRNA-derived sncRNAs should include review of studies looking into the characteristics of ssRNA ligands of TLR7 and TLR8.
Beyond tRNA-derived sncRNAs, our first genome-wide identification of cP-containing sncRNAs revealed abundant expression of rRNA- and mRNA-derived sncRNAs in various tissues (37), and, indeed, not only mature rRNAs (79) but also rRNA- and mRNA-derived sncRNAs have been shown to be abundantly secreted as ex-RNAs (26,80,81). These other unexplored classes of ex-sncRNAs should further be investigated as potential activators of TLR7 and TLR8.
Endosomal TLR3 also recognizes RNA molecules, but it binds to double-stranded RNAs (dsRNAs) (82). Although ssRNAs containing partial stem structures can also be TLR3 ligands, at least 40–50-base-pairs (bp) length of dsRNA molecule is required to ensure stable complex formation with TLR3 (83,84), and more than 90-bp length of dsRNA is required to activate TLR3 signal transduction in dendritic cell maturation (85). Therefore, ex-sncRNAs shorter than mature tRNAs are unlikely to participate in the TLR3 signaling. However, exRNAs more broadly should be considered as potentially major endogenous ligands of TLR3, as exemplified in a study showing that dsRNAs released from necrotic polymorphonuclear neutrophils induce inflammatory response in macrophages (86).
Although mature tRNAs have been reported to be incorporated into EVs (70,87), unlike the 5'-tRNAHisGUG half, the full-length tRNAHisGUG was incapable of stimulating endosomal TLR (20), possibly due to its rigid secondary and tertiary structures. Therefore, shortening mature tRNAHisGUG into less-rigid 5'-half molecules by anticodon-cleavage is crucial to produce immunostimulatory sncRNAs. ANG is highly enriched in the EVs derived from aggressive brain tumor cells (52), while another RNase A superfamily member, RNase 1, has recently been shown to cleave non-vesicular tRNAs (88). Further research on immune responsive ribonucleases which cleave tRNAs, rRNAs, mRNAs, or other substrate RNAs to produce immune responsive ex-sncRNAs is necessary to fully understand the regulation and functional consequences of immune response mediated by these ex-sncRNAs.
TLR7 and TLR8 are involved in various biological processes and diseases (89). The symptom severity of COVID-19 is associated with TLR7 mutations (90-92). The genetic polymorphisms in TLR7 and TLR8 genes, which reduce the TLR activity, increase susceptibility to Mtb infection (93), and upregulation and stimulation of TLR7 in macrophages suppress Mtb growth (94,95). TLR7 and TLR8 have been implicated in the progression of Parkinson’s disease (96) and in Alzheimer’s disease (12,97,98), possibly playing a role in promoting neuroinflammation and autoimmunity. A recent study also linked a specific gain-of-function mutation in TLR7 with severe systemic lupus erythematosus (99). Furthermore, missense mutations in TLR7 have been reported in the tumors of a subset of esophageal adenocarcinoma patients (100), and cellular immune response through activation of TLR7 and TLR8 can be directed against tumors (101). Despite these known linkages between TLR7/TLR8 and pathobiology of diseases, the endogenous ligands of these ssRNA-sensing TLRs remain poorly understood. The growing evidence of TLR7 and TLR8 involvement in the pathogenesis of non-infectious diseases highlights the need for a better understanding of the endogenous ligands of these receptors. Further research into tRNA-derived ex-sncRNAs, aided by upgraded sequencing methods, is necessary to identify these ligands and to gain a fuller understanding of the interplay between ex-RNAs, TLRs, and the immune system in the body’s response to diverse pathologies.
Acknowledgments
We are grateful to Megumi Shigematsu and Takuya Kawamura (Thomas Jefferson University) for helpful discussions.
Funding: Work in the Kirino lab on this topic is currently supported by the National Institutes of Health Grants (Nos. GM106047, HL150560, AI151641, and AI168975 to YK) and the American Cancer Society Research Scholar Grant (No. RSG-17-059-01-RMC to YK).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Silvano Sozzani and Daniela Bosisio) for the series “Extracellular MiRNAs as Activators of Innate Immune Receptors” published in ExRNA. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-22/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-22/coif). The series “Extracellular MiRNAs as Activators of Innate Immune Receptors” was commissioned by the editorial office without any funding or sponsorship. YK reports funding from National Institutes of Health Grants (Nos. GM106047, HL150560, AI151641, and AI168975) and the American Cancer Society Research Scholar Grant (No. RSG-17-059-01-RMC). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brubaker SW, Bonham KS, Zanoni I, et al. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015;33:257-90. [Crossref] [PubMed]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373-84. [Crossref] [PubMed]
- Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol 2014;5:461. [Crossref] [PubMed]
- Satoh T, Akira S. Toll-Like Receptor Signaling and Its Inducible Proteins. Microbiol Spectr 2016;4: [Crossref] [PubMed]
- Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004;303:1526-9. [Crossref] [PubMed]
- Zhang Z, Ohto U, Shibata T, et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016;45:737-48. [Crossref] [PubMed]
- Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 2004;101:5598-603. [Crossref] [PubMed]
- Melchjorsen J, Jensen SB, Malmgaard L, et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol 2005;79:12944-51. [Crossref] [PubMed]
- Triantafilou K, Orthopoulos G, Vakakis E, et al. Human cardiac inflammatory responses triggered by Coxsackie B viruses are mainly Toll-like receptor (TLR) 8-dependent. Cell Microbiol 2005;7:1117-26. [Crossref] [PubMed]
- Gantier MP, Irving AT, Kaparakis-Liaskos M, et al. Genetic modulation of TLR8 response following bacterial phagocytosis. Hum Mutat 2010;31:1069-79. [Crossref] [PubMed]
- Rifkin IR, Leadbetter EA, Busconi L, et al. Toll-like receptors, endogenous ligands, and systemic autoimmune disease. Immunol Rev 2005;204:27-42. [Crossref] [PubMed]
- Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci 2012;15:827-35. [Crossref] [PubMed]
- Fabbri M, Paone A, Calore F, et al. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol 2013;10:169-74. [Crossref] [PubMed]
- Feng Y, Zou L, Yan D, et al. Extracellular MicroRNAs Induce Potent Innate Immune Responses via TLR7/MyD88-Dependent Mechanisms. J Immunol 2017;199:2106-17. [Crossref] [PubMed]
- Zou L, He J, Gu L, et al. Brain innate immune response via miRNA-TLR7 sensing in polymicrobial sepsis. Brain Behav Immun 2022;100:10-24. [Crossref] [PubMed]
- Fabbri M. TLRs as miRNA receptors. Cancer Res 2012;72:6333-7. [Crossref] [PubMed]
- Casadei L, Calore F, Creighton CJ, et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res 2017;77:3846-56. [Crossref] [PubMed]
- Wu WC, Song SJ, Zhang Y, et al. Role of Extracellular Vesicles in Autoimmune Pathogenesis. Front Immunol 2020;11:579043. [Crossref] [PubMed]
- Groot M, Lee H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020;9:1044. [Crossref] [PubMed]
- Pawar K, Shigematsu M, Sharbati S, et al. Infection-induced 5'-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol 2020;18:e3000982. [Crossref] [PubMed]
- Shigematsu M, Kirino Y. Making Invisible RNA Visible: Discriminative Sequencing Methods for RNA Molecules with Specific Terminal Formations. Biomolecules 2022;12:611. [Crossref] [PubMed]
- Balka KR, De Nardo D. Understanding early TLR signaling through the Myddosome. J Leukoc Biol 2019;105:339-51. [Crossref] [PubMed]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 2008;8:594-606. [Crossref] [PubMed]
- Shigematsu M, Kawamura T, Kirino Y. Generation of 2',3'-Cyclic Phosphate-Containing RNAs as a Hidden Layer of the Transcriptome. Front Genet 2018;9:562. [Crossref] [PubMed]
- Giraldez MD, Spengler RM, Etheridge A, et al. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J 2019;38:e101695. [Crossref] [PubMed]
- Akat KM, Lee YA, Hurley A, et al. Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight 2019;5:e127317. [Crossref] [PubMed]
- Qin Y, Yao J, Wu DC, et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016;22:111-28. [Crossref] [PubMed]
- Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci 2016;41:679-89. [Crossref] [PubMed]
- Shigematsu M, Kirino Y. tRNA-Derived Short Non-coding RNA as Interacting Partners of Argonaute Proteins. Gene Regul Syst Bio 2015;9:27-33. [Crossref] [PubMed]
- Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett 2014;588:4297-304. [Crossref] [PubMed]
- Magee R, Rigoutsos I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res 2020;48:9433-48. [Crossref] [PubMed]
- Yamasaki S, Ivanov P, Hu GF, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009;185:35-42. [Crossref] [PubMed]
- Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 2009;583:437-42. [Crossref] [PubMed]
- Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry 1986;25:3527-32. [Crossref] [PubMed]
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A 2015;112:E3816-25. [Crossref] [PubMed]
- Honda S, Kawamura T, Loher P, et al. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res 2017;45:9108-20. [Crossref] [PubMed]
- Shigematsu M, Morichika K, Kawamura T, et al. Genome-wide identification of short 2',3'-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet 2019;15:e1008469. [Crossref] [PubMed]
- Shigematsu M, Kirino Y. Oxidative stress enhances the expression of 2',3'-cyclic phosphate-containing RNAs. RNA Biol 2020;17:1060-9. [Crossref] [PubMed]
- Shigematsu M, Kawamura T, Morichika K, et al. RNase κ promotes robust piRNA production by generating 2',3'-cyclic phosphate-containing precursors. Nat Commun 2021;12:4498. [Crossref] [PubMed]
- Saikia M, Hatzoglou M. The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health. J Biol Chem 2015;290:29761-8. [Crossref] [PubMed]
- Saikia M, Krokowski D, Guan BJ, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 2012;287:42708-25. [Crossref] [PubMed]
- Pizzo E, Sarcinelli C, Sheng J, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci 2013;126:4308-19. [Crossref] [PubMed]
- Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010;285:10959-68. [Crossref] [PubMed]
- Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011;43:613-23. [Crossref] [PubMed]
- Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014;33:2020-39. [Crossref] [PubMed]
- Hogg MC, Rayner M, Susdalzew S, et al. 5'ValCAC tRNA fragment generated as part of a protective angiogenin response provides prognostic value in amyotrophic lateral sclerosis. Brain Commun 2020;2:fcaa138. [Crossref] [PubMed]
- Haussecker D, Huang Y, Lau A, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010;16:673-95. [Crossref] [PubMed]
- Li Z, Ender C, Meister G, et al. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012;40:6787-99. [Crossref] [PubMed]
- Yu X, Xie Y, Zhang S, et al. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021;11:461-9. [Crossref] [PubMed]
- Nolte-'t Hoen EN, Buermans HP, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res 2012;40:9272-85. [Crossref] [PubMed]
- Baglio SR, Rooijers K, Koppers-Lalic D, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther 2015;6:127. [Crossref] [PubMed]
- Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun 2017;8:1145. [Crossref] [PubMed]
- Tosar JP, Gámbaro F, Sanguinetti J, et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res 2015;43:5601-16. [Crossref] [PubMed]
- Allen RM, Zhao S, Ramirez Solano MA, et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J Extracell Vesicles 2018;7:1506198. [Crossref] [PubMed]
- Sharma U, Sun F, Conine CC, et al. Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev Cell 2018;46:481-494.e6. [Crossref] [PubMed]
- Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391-6. [Crossref] [PubMed]
- Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397-400. [Crossref] [PubMed]
- Dhahbi JM, Spindler SR, Atamna H, et al. 5' tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 2013;14:298. [Crossref] [PubMed]
- Dhahbi JM, Spindler SR, Atamna H, et al. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5' tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark Cancer 2014;6:37-47. [Crossref] [PubMed]
- Dhahbi JM, Atamna H, Selth LA. Data Mining of Small RNA-Seq Suggests an Association Between Prostate Cancer and Altered Abundance of 5' Transfer RNA Halves in Seminal Fluid and Prostatic Tissues. Biomark Cancer 2018;10:1179299X18759545.
- Shao Y, Sun Q, Liu X, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des 2017;90:730-8. [Crossref] [PubMed]
- Khurana R, Ranches G, Schafferer S, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA 2017;23:142-52. [Crossref] [PubMed]
- Tosar JP, Cayota A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol 2020;17:1149-67. [Crossref] [PubMed]
- Wang J, Ma G, Li M, et al. Plasma tRNA Fragments Derived from 5' Ends as Novel Diagnostic Biomarkers for Early-Stage Breast Cancer. Mol Ther Nucleic Acids 2020;21:954-64. [Crossref] [PubMed]
- Koi Y, Tsutani Y, Nishiyama Y, et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci 2020;111:2104-15. [Crossref] [PubMed]
- Wang J, Ma G, Ge H, et al. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ Breast Cancer 2021;7:4. [Crossref] [PubMed]
- Xi J, Zeng Z, Li X, et al. Expression and Diagnostic Value of tRNA-Derived Fragments Secreted by Extracellular Vesicles in Hypopharyngeal Carcinoma. Onco Targets Ther 2021;14:4189-99. [Crossref] [PubMed]
- Yang P, Zhang X, Chen S, et al. A Novel Serum tsRNA for Diagnosis and Prediction of Nephritis in SLE. Front Immunol 2021;12:735105. [Crossref] [PubMed]
- Lee HK, Lee BR, Lee TJ, et al. Differential release of extracellular vesicle tRNA from oxidative stressed renal cells and ischemic kidneys. Sci Rep 2022;12:1646. [Crossref] [PubMed]
- Shurtleff MJ, Yao J, Qin Y, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A 2017;114:E8987-95. [Crossref] [PubMed]
- Kawamura T, Shigematsu M, Kirino Y. In vitro production and multiplex quantification of 2',3'-cyclic phosphate-containing 5'-tRNA half molecules. Methods 2022;203:335-41. [Crossref] [PubMed]
- Zhang Y, Zhang Y, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol 2014;6:172-4. [Crossref] [PubMed]
- Zhou J, Liu S, Chen Y, et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol 2017;98:1600-10. [Crossref] [PubMed]
- Deng J, Ptashkin RN, Chen Y, et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol Ther 2015;23:1622-9. [Crossref] [PubMed]
- Gong B, Lee YS, Lee I, et al. Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect Dis 2013;13:285. [Crossref] [PubMed]
- Selitsky SR, Baran-Gale J, Honda M, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 2015;5:7675. [Crossref] [PubMed]
- Tanji H, Ohto U, Shibata T, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat Struct Mol Biol 2015;22:109-15. [Crossref] [PubMed]
- Greulich W, Wagner M, Gaidt MM, et al. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019;179:1264-1275.e13. [Crossref] [PubMed]
- Preissner KT, Fischer S. Functions and cellular signaling by ribosomal extracellular RNA (rexRNA): Facts and hypotheses on a non-typical DAMP. Biochim Biophys Acta Mol Cell Res 2023;1870:119408. [Crossref] [PubMed]
- Li M, Zeringer E, Barta T, et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci 2014;369:20130502. [Crossref] [PubMed]
- Jia J, Yang S, Huang J, et al. Distinct Extracellular RNA Profiles in Different Plasma Components. Front Genet 2021;12:564780. [Crossref] [PubMed]
- Alexopoulou L, Holt AC, Medzhitov R, et al. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001;413:732-8. [Crossref] [PubMed]
- Liu L, Botos I, Wang Y, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008;320:379-81. [Crossref] [PubMed]
- Leonard JN, Ghirlando R, Askins J, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A 2008;105:258-63. [Crossref] [PubMed]
- Jelinek I, Leonard JN, Price GE, et al. TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J Immunol 2011;186:2422-9. [Crossref] [PubMed]
- Cavassani KA, Ishii M, Wen H, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med 2008;205:2609-21. [Crossref] [PubMed]
- Tosar JP, Segovia M, Castellano M, et al. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res 2020;48:12874-88. [Crossref] [PubMed]
- Nechooshtan G, Yunusov D, Chang K, et al. Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res 2020;48:8035-49. [Crossref] [PubMed]
- Lind NA, Rael VE, Pestal K, et al. Regulation of the nucleic acid-sensing Toll-like receptors. Nat Rev Immunol 2022;22:224-35. [Crossref] [PubMed]
- Asano T, Boisson B, Onodi F, et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci Immunol 2021;6:eabl4348. [Crossref] [PubMed]
- Solanich X, Vargas-Parra G, van der Made CI, et al. Genetic Screening for TLR7 Variants in Young and Previously Healthy Men With Severe COVID-19. Front Immunol 2021;12:719115. [Crossref] [PubMed]
- Fallerini C, Daga S, Mantovani S, et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife 2021;10:e67569. [Crossref] [PubMed]
- Lai YF, Lin TM, Wang CH, et al. Functional polymorphisms of the TLR7 and TLR8 genes contribute to Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2016;98:125-31. [Crossref] [PubMed]
- Bao M, Yi Z, Fu Y. Activation of TLR7 Inhibition of Mycobacterium Tuberculosis Survival by Autophagy in RAW 264.7 Macrophages. J Cell Biochem 2017;118:4222-9. [Crossref] [PubMed]
- Lee HJ, Kang SJ, Woo Y, et al. TLR7 Stimulation With Imiquimod Induces Selective Autophagy and Controls Mycobacterium tuberculosis Growth in Mouse Macrophages. Front Microbiol 2020;11:1684. [Crossref] [PubMed]
- Campolo M, Filippone A, Biondo C, et al. TLR7/8 in the Pathogenesis of Parkinson's Disease. Int J Mol Sci 2020;21:9384. [Crossref] [PubMed]
- Frank S, Copanaki E, Burbach GJ, et al. Differential regulation of toll-like receptor mRNAs in amyloid plaque-associated brain tissue of aged APP23 transgenic mice. Neurosci Lett 2009;453:41-4. [Crossref] [PubMed]
- Dembny P, Newman AG, Singh M, et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight 2020;5:e131093. [Crossref] [PubMed]
- Brown GJ, Cañete PF, Wang H, et al. TLR7 gain-of-function genetic variation causes human lupus. Nature 2022;605:349-56. [Crossref] [PubMed]
- Fels Elliott DR, Perner J, Li X, et al. Impact of mutations in Toll-like receptor pathway genes on esophageal carcinogenesis. PLoS Genet 2017;13:e1006808. [Crossref] [PubMed]
- Schön MP, Schön M. TLR7 and TLR8 as targets in cancer therapy. Oncogene 2008;27:190-9. [Crossref] [PubMed]
Cite this article as: Gumas J, Kirino Y. Extracellular tRNA-derived RNAs as emerging activators of endosomal Toll-like receptors: a narrative review. ExRNA 2023;5:2.


